Chromosome organization by a conserved condensin-ParB system in the actinobacterium Corynebacterium glutamicum
- PMID: 32198399
- PMCID: PMC7083940
- DOI: 10.1038/s41467-020-15238-4
Chromosome organization by a conserved condensin-ParB system in the actinobacterium Corynebacterium glutamicum
Abstract
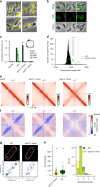
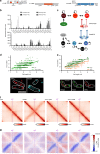
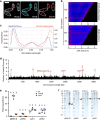
Higher-order chromosome folding and segregation are tightly regulated in all domains of life. In bacteria, details on nucleoid organization regulatory mechanisms and function remain poorly characterized, especially in non-model species. Here, we investigate the role of DNA-partitioning protein ParB and SMC condensin complexes in the actinobacterium Corynebacterium glutamicum. Chromosome conformation capture reveals SMC-mediated long-range interactions around ten centromere-like parS sites clustered at the replication origin (oriC). At least one oriC-proximal parS site is necessary for reliable chromosome segregation. We use chromatin immunoprecipitation and photoactivated single-molecule localization microscopy to show the formation of distinct, parS-dependent ParB-nucleoprotein subclusters. We further show that SMC/ScpAB complexes, loaded via ParB at parS sites, mediate chromosomal inter-arm contacts (as previously shown in Bacillus subtilis). However, the MukBEF-like SMC complex MksBEFG does not contribute to chromosomal DNA-folding; instead, this complex is involved in plasmid maintenance and interacts with the polar oriC-tethering factor DivIVA. Our results complement current models of ParB-SMC/ScpAB crosstalk and show that some condensin complexes evolved functions that are apparently uncoupled from chromosome folding.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

