The genetic legacy of extreme exploitation in a polar vertebrate
- PMID: 32198403
- PMCID: PMC7083876
- DOI: 10.1038/s41598-020-61560-8
The genetic legacy of extreme exploitation in a polar vertebrate
Erratum in
-
Publisher Correction: The genetic legacy of extreme exploitation in a polar vertebrate.Sci Rep. 2020 May 12;10(1):8098. doi: 10.1038/s41598-020-64763-1. Sci Rep. 2020. PMID: 32398874 Free PMC article.
Abstract
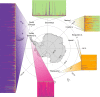
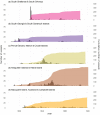
Understanding the effects of human exploitation on the genetic composition of wild populations is important for predicting species persistence and adaptive potential. We therefore investigated the genetic legacy of large-scale commercial harvesting by reconstructing, on a global scale, the recent demographic history of the Antarctic fur seal (Arctocephalus gazella), a species that was hunted to the brink of extinction by 18th and 19th century sealers. Molecular genetic data from over 2,000 individuals sampled from all eight major breeding locations across the species' circumpolar geographic distribution, show that at least four relict populations around Antarctica survived commercial hunting. Coalescent simulations suggest that all of these populations experienced severe bottlenecks down to effective population sizes of around 150-200. Nevertheless, comparably high levels of neutral genetic variability were retained as these declines are unlikely to have been strong enough to deplete allelic richness by more than around 15%. These findings suggest that even dramatic short-term declines need not necessarily result in major losses of diversity, and explain the apparent contradiction between the high genetic diversity of this species and its extreme exploitation history.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Erickson JD. Endangering the economics of extinction. Wildl. Soc. Bull. 2000;28:34–41.
-
- Peres, C. A. Overharvesting. In Conservation biology for all (eds. Sodhi, N. S. & Ehrlich, P. R.) (Oxford University Press, 2010).
-
- IUCN red list of threatened species: a global species assessment. (IUCN, 2004). 2004
-
- Farrow S. Extinction and market forces: two case studies. Ecol. Econ. 1995;13:115–123. doi: 10.1016/0921-8009(94)00075-7. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

