Enhanced purification coupled with biophysical analyses shows cross-β structure as a core building block for Streptococcus mutans functional amyloids
- PMID: 32198417
- PMCID: PMC7083922
- DOI: 10.1038/s41598-020-62115-7
Enhanced purification coupled with biophysical analyses shows cross-β structure as a core building block for Streptococcus mutans functional amyloids
Abstract
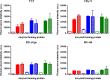
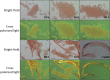
Streptococcus mutans is an etiologic agent of human dental caries that forms dental plaque biofilms containing functional amyloids. Three amyloidogenic proteins, P1, WapA, and Smu_63c were previously identified. C123 and AgA are naturally occurring amyloid-forming fragments of P1 and WapA, respectively. We determined that four amyloidophilic dyes, ThT, CDy11, BD-oligo, and MK-H4, differentiate C123, AgA, and Smu_63c amyloid from monomers, but non-specific binding to bacterial cells in the absence of amyloid precludes their utility for identifying amyloid in biofilms. Congo red-induced birefringence is a more specific indicator of amyloid formation and differentiates biofilms formed by wild-type S. mutans from a triple ΔP1/WapA/Smu_63c mutant with reduced biofilm forming capabilities. Amyloid accumulation is a late event, appearing in older S. mutans biofilms after 60 hours of growth. Amyloid derived from pure preparations of all three proteins is visualized by electron microscopy as mat-like structures. Typical amyloid fibers become evident following protease digestion to eliminate non-specific aggregates and monomers. Amyloid mats, similar in appearance to those reported in S. mutans biofilm extracellular matrices, are reconstituted by co-incubation of monomers and amyloid fibers. X-ray fiber diffraction of amyloid mats and fibers from all three proteins demonstrate patterns reflective of a cross-β amyloid structure.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Functional amyloids in Streptococcus mutans, their use as targets of biofilm inhibition and initial characterization of SMU_63c.Microbiology (Reading). 2017 Apr;163(4):488-501. doi: 10.1099/mic.0.000443. Epub 2017 Apr 26. Microbiology (Reading). 2017. PMID: 28141493 Free PMC article.
-
Amyloid Aggregates Are Localized to the Nonadherent Detached Fraction of Aging Streptococcus mutans Biofilms.Microbiol Spectr. 2022 Aug 31;10(4):e0166122. doi: 10.1128/spectrum.01661-22. Epub 2022 Aug 11. Microbiol Spectr. 2022. PMID: 35950854 Free PMC article.
-
Functional amyloid formation by Streptococcus mutans.Microbiology (Reading). 2012 Dec;158(Pt 12):2903-2916. doi: 10.1099/mic.0.060855-0. Epub 2012 Oct 18. Microbiology (Reading). 2012. PMID: 23082034 Free PMC article.
-
Strategies for Streptococcus mutans biofilm dispersal through extracellular polymeric substances disruption.Mol Oral Microbiol. 2022 Feb;37(1):1-8. doi: 10.1111/omi.12355. Epub 2021 Nov 22. Mol Oral Microbiol. 2022. PMID: 34727414 Review.
-
Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms.Caries Res. 2011;45(1):69-86. doi: 10.1159/000324598. Epub 2011 Feb 23. Caries Res. 2011. PMID: 21346355 Free PMC article. Review.
Cited by
-
The Group B Streptococcal Adhesin BspC Interacts with Host Cytokeratin 19 To Promote Colonization of the Female Reproductive Tract.mBio. 2022 Oct 26;13(5):e0178122. doi: 10.1128/mbio.01781-22. Epub 2022 Sep 7. mBio. 2022. PMID: 36069447 Free PMC article.
-
Distinct Agents Induce Streptococcus mutans Cells with Altered Biofilm Formation Capacity.Microbiol Spectr. 2022 Aug 31;10(4):e0065022. doi: 10.1128/spectrum.00650-22. Epub 2022 Jul 11. Microbiol Spectr. 2022. PMID: 35862994 Free PMC article.
-
Synchrotron X-ray Studies of the Structural and Functional Hierarchies in Mineralised Human Dental Enamel: A State-of-the-Art Review.Dent J (Basel). 2023 Apr 7;11(4):98. doi: 10.3390/dj11040098. Dent J (Basel). 2023. PMID: 37185477 Free PMC article. Review.
-
Biological Functions of Prokaryotic Amyloids in Interspecies Interactions: Facts and Assumptions.Int J Mol Sci. 2020 Sep 30;21(19):7240. doi: 10.3390/ijms21197240. Int J Mol Sci. 2020. PMID: 33008049 Free PMC article. Review.
-
Amyloid Fibrils Produced by Streptococcus sanguinis Contribute to Biofilm Formation and Immune Evasion.Int J Mol Sci. 2023 Oct 28;24(21):15686. doi: 10.3390/ijms242115686. Int J Mol Sci. 2023. PMID: 37958670 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

