Small circRNAs with self-cleaving ribozymes are highly expressed in diverse metazoan transcriptomes
- PMID: 32198887
- PMCID: PMC7229834
- DOI: 10.1093/nar/gkaa187
Small circRNAs with self-cleaving ribozymes are highly expressed in diverse metazoan transcriptomes
Abstract
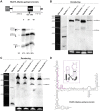
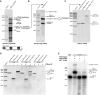
Ribozymes are catalytic RNAs present in modern genomes but regarded as remnants of a prebiotic RNA world. The paradigmatic hammerhead ribozyme (HHR) is a small self-cleaving motif widespread from bacterial to human genomes. Here, we report that most of the classical type I HHRs frequently found in the genomes of animals are contained within a novel family of non-autonomous non-LTR retrotransposons of the retrozyme class. These retroelements are expressed as abundant linear and circular RNAs of ∼170-400 nt in different animal tissues. Bioinformatic and in vitro analyses indicate an efficient self-cleavage of the HHRs harboured in most invertebrate retrozymes, whereas HHRs in retrozymes of vertebrates, such as the axolotl and other amphibians, require to act as dimeric motifs to reach higher self-cleavage rates. Ligation assays of retrozyme RNAs with a protein ligase versus HHR self-ligation indicate that, most likely, tRNA ligases and not the ribozymes are involved in the step of RNA circularization. Altogether, these results confirm the existence of a new and conserved pathway in animals and, likely, eukaryotes in general, for the efficient biosynthesis of RNA circles through small ribozymes, which opens the door for the development of new tools in the emerging field of study of circRNAs.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R.. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982; 31:147–157. - PubMed
-
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:849–857. - PubMed
-
- Gilbert W. The RNA world. Nature. 1986; 319:618.
-
- Orgel L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968; 38:381–393. - PubMed
-
- Crick F.H. The origin of the genetic code. J. Mol. Biol. 1968; 38:367–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

