Drivers and regulators of humoral innate immune responses to infection and cancer
- PMID: 32199212
- PMCID: PMC7207242
- DOI: 10.1016/j.molimm.2020.03.005
Drivers and regulators of humoral innate immune responses to infection and cancer
Abstract
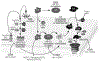
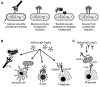
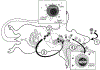
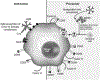
The complement cascade consists of cell bound and serum proteins acting together to protect the host from pathogens, remove cancerous cells and effectively links innate and adaptive immune responses. Despite its usefulness in microbial neutralization and clearance of cancerous cells, excessive complement activation causes an immune imbalance and tissue damage in the host. Hence, a series of complement regulatory proteins present at a higher concentration in blood plasma and on cell surfaces tightly regulate the cascade. The complement cascade can be initiated by B-1 B cell production of natural antibodies. Natural antibodies arise spontaneously without any known exogenous antigenic or microbial stimulus and protect against invading pathogens, clear apoptotic cells, provide tissue homeostasis, and modulate adaptive immune functions. Natural IgM antibodies recognize microbial and cancer antigens and serve as an activator of complement mediated lysis. This review will discuss advances in complement activation and regulation in bacterial and viral infections, and cancer. We will also explore the crosstalk of natural antibodies with bacterial populations and cancer.
Keywords: B-1 B cells; Bacteria; Cancer; Complement; Natural antibodies; Virus.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Role of natural and immune IgM antibodies in immune responses.Mol Immunol. 2000 Dec;37(18):1141-9. doi: 10.1016/s0161-5890(01)00025-6. Mol Immunol. 2000. PMID: 11451419 Review.
-
Evasion and interactions of the humoral innate immune response in pathogen invasion, autoimmune disease, and cancer.Clin Immunol. 2015 Oct;160(2):244-54. doi: 10.1016/j.clim.2015.06.012. Epub 2015 Jul 2. Clin Immunol. 2015. PMID: 26145788 Free PMC article. Review.
-
Interactions of viruses and the humoral innate immune response.Clin Immunol. 2020 Mar;212:108351. doi: 10.1016/j.clim.2020.108351. Epub 2020 Feb 4. Clin Immunol. 2020. PMID: 32028020 Free PMC article. Review.
-
Dendritic cell development in infection.Mol Immunol. 2020 May;121:111-117. doi: 10.1016/j.molimm.2020.02.015. Epub 2020 Mar 18. Mol Immunol. 2020. PMID: 32199210 Review.
-
Complement: an efficient sword of innate immunity.Contrib Microbiol. 2008;15:78-100. doi: 10.1159/000136316. Contrib Microbiol. 2008. PMID: 18511857 Review.
Cited by
-
The Forgotten Brother: The Innate-like B1 Cell in Multiple Sclerosis.Biomedicines. 2022 Mar 4;10(3):606. doi: 10.3390/biomedicines10030606. Biomedicines. 2022. PMID: 35327408 Free PMC article. Review.
-
Plant-Derived Nutraceuticals and Immune System Modulation: An Evidence-Based Overview.Vaccines (Basel). 2020 Aug 22;8(3):468. doi: 10.3390/vaccines8030468. Vaccines (Basel). 2020. PMID: 32842641 Free PMC article. Review.
-
Delivery of a novel membrane-anchored Fc chimera enhances NK cell-mediated killing of tumor cells and persistently virus-infected cells.PLoS One. 2023 May 5;18(5):e0285532. doi: 10.1371/journal.pone.0285532. eCollection 2023. PLoS One. 2023. PMID: 37146009 Free PMC article.
-
Sialic Acid Protects Nontypeable Haemophilus influenzae from Natural IgM and Promotes Survival in Murine Respiratory Tract.Infect Immun. 2021 May 17;89(6):e00676-20. doi: 10.1128/IAI.00676-20. Print 2021 May 17. Infect Immun. 2021. PMID: 33782153 Free PMC article.
-
How Children Are Protected From COVID-19? A Historical, Clinical, and Pathophysiological Approach to Address COVID-19 Susceptibility.Front Immunol. 2021 Jun 11;12:646894. doi: 10.3389/fimmu.2021.646894. eCollection 2021. Front Immunol. 2021. PMID: 34177895 Free PMC article.
References
-
- Ajona D, Castano Z, Garayoa M, Zudaire E, Pajares MJ, Martinez A, Cuttitta F, Montuenga LM, Pio R, 2004. Expression of complement factor H by lung cancer cells: effects on the activation of the alternative pathway of complement. Cancer Res 64, 6310–6318. 10.1158/0008-5472.can-03-2328. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

