Interactions between metabolism and chromatin in plant models
- PMID: 32199818
- PMCID: PMC7300381
- DOI: 10.1016/j.molmet.2020.01.015
Interactions between metabolism and chromatin in plant models
Erratum in
-
Corrigendum to "Interactions between metabolism and chromatin in plant models" [Mol Metab 38 (Aug 2020) 100951].Mol Metab. 2020 Nov;41:101069. doi: 10.1016/j.molmet.2020.101069. Epub 2020 Sep 4. Mol Metab. 2020. PMID: 32891669 Free PMC article. No abstract available.
Abstract
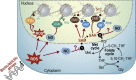
Background: One of the fascinating aspects of epigenetic regulation is that it provides means to rapidly adapt to environmental change. This is particularly relevant in the plant kingdom, where most species are sessile and exposed to increasing habitat fluctuations due to global warming. Although the inheritance of epigenetically controlled traits acquired through environmental impact is a matter of debate, it is well documented that environmental cues lead to epigenetic changes, including chromatin modifications, that affect cell differentiation or are associated with plant acclimation and defense priming. Still, in most cases, the mechanisms involved are poorly understood. An emerging topic that promises to reveal new insights is the interaction between epigenetics and metabolism.
Scope of review: This study reviews the links between metabolism and chromatin modification, in particular histone acetylation, histone methylation, and DNA methylation, in plants and compares them to examples from the mammalian field, where the relationship to human diseases has already generated a larger body of literature. This study particularly focuses on the role of reactive oxygen species (ROS) and nitric oxide (NO) in modulating metabolic pathways and gene activities that are involved in these chromatin modifications. As ROS and NO are hallmarks of stress responses, we predict that they are also pivotal in mediating chromatin dynamics during environmental responses.
Major conclusions: Due to conservation of chromatin-modifying mechanisms, mammals and plants share a common dependence on metabolic intermediates that serve as cofactors for chromatin modifications. In addition, plant-specific non-CG methylation pathways are particularly sensitive to changes in folate-mediated one-carbon metabolism. Finally, reactive oxygen and nitrogen species may fine-tune epigenetic processes and include similar signaling mechanisms involved in environmental stress responses in plants as well as animals.
Keywords: Chromatin; DNA methylation; Folate metabolism; Histone modification; Methionine cycle; Nitric oxide; Plants; Reactive oxygen species; Redox modification.
Copyright © 2020 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures
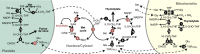
References
-
- Hartmann T. From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry. 2007;68:2831–2846. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

