Transcriptomic analysis reveals the oncogenic role of S6K1 in hepatocellular carcinoma
- PMID: 32201535
- PMCID: PMC7065997
- DOI: 10.7150/jca.40726
Transcriptomic analysis reveals the oncogenic role of S6K1 in hepatocellular carcinoma
Abstract
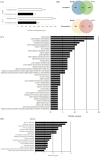
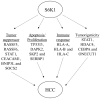
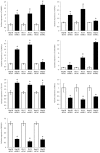
The p70 ribosomal protein S6 kinase 1 (S6K1), a serine/threonine kinase, is commonly overexpressed in a variety of cancers. However, its expression level and functional roles in hepatocellular carcinoma (HCC), which ranks as the third leading cause of cancer-related death worldwide, is still largely unknown. In the current report, we show the in vivo and in vitro overexpression of S6K1 in HCC. In the functional analysis, we demonstrate that S6K1 is required for the proliferation and colony formation abilities in HCC. By using comparative transcriptomic analysis followed by gene ontology enrichment analysis and Ingenuity Pathway Analysis, we find that the depletion of S6K1 can elevate the expression of a cluster of apoptotic genes, tumor suppressor genes and immune responsive genes. Moreover, the knockdown of S6K1 is predicted to reduce the tumorigenicity of HCC through the regulation of hubs of genes including STAT1, HDAC4, CEBPA and ONECUT1. In conclusion, we demonstrate the oncogenic role of S6K1 in HCC, suggesting the possible use of S6K1 as a therapeutic target for HCC treatment.
Keywords: S6K1; hepatocellular carcinoma; proliferation; transcriptome; tumorigenicity.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Giannelli G, Rani B, Dituri F. et al. Moving towards personalised therapy in patients with hepatocellular carcinoma: the role of the microenvironment. Gut. 2014;63:1668–1676. - PubMed
-
- Lai KP, Chen J, He M. et al. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014;135:1790–1799. - PubMed
-
- Liu L, Dai Y, Chen J. et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial-mesenchymal transition by way of Akt/GSK-3beta/Snail signaling. Hepatology. 2014;59:531–543. - PubMed
-
- Ma S, Lee TK, Zheng BJ. et al. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–1758. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

