Differences in Endothelin B Receptor Isoforms Expression and Function in Breast Cancer Cells
- PMID: 32201539
- PMCID: PMC7066022
- DOI: 10.7150/jca.41004
Differences in Endothelin B Receptor Isoforms Expression and Function in Breast Cancer Cells
Abstract
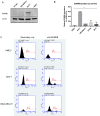
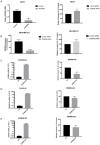
The endothelins and their receptors are best known for their regulation of the vascular system. Their widespread expression in epithelial cells and their overexpression in some tumors has prompted investigation into their ability to regulate cancer progression. In this study, we assessed the mRNA expression of the major endothelin B receptor gene (EDNRB) isoforms and found differences in both mRNA and protein expression in normal breast cells and breast cancer cell lines. Knocking down the EDNRB gene in breast cancer cells altered invasiveness toward endothelin 3 (ET3), and we observed EDNRB isoform-specific regulation of breast cancer cell invasion and cell signaling, as well as isoform- and subtype-specific differences in breast cancer patient survival. The results reported in this study emphasize the importance of the endothelin B receptor in breast cancer. To our knowledge, this study is the first to clarify the differential expression and roles of specific EDNRB isoforms in breast cancer.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Knockdown of endothelin receptor B inhibits the progression of triple-negative breast cancer.Ann N Y Acad Sci. 2019 Jul;1448(1):5-18. doi: 10.1111/nyas.14039. Epub 2019 Mar 22. Ann N Y Acad Sci. 2019. PMID: 30900271
-
Hypoxic regulation of EDN1, EDNRA, EDNRB, and ECE1 gene expressions in ERN1 knockdown U87 glioma cells.Endocr Regul. 2019 Oct 1;53(4):250-262. doi: 10.2478/enr-2019-0025. Endocr Regul. 2019. PMID: 31734650
-
Altered endothelin-3 and endothelin-B receptor mRNA expression in Hirschsprung's disease.J Pediatr Surg. 1999 Aug;34(8):1257-60. doi: 10.1016/s0022-3468(99)90163-x. J Pediatr Surg. 1999. PMID: 10466607
-
Endothelins in breast tumour cell invasion.Cancer Lett. 2005 May 26;222(2):129-38. doi: 10.1016/j.canlet.2004.08.029. Cancer Lett. 2005. PMID: 15863261 Review.
-
Endothelin receptor B is required for the expansion of melanocyte precursors and malignant melanoma.Int J Dev Biol. 2005;49(2-3):173-80. doi: 10.1387/ijdb.041951rl. Int J Dev Biol. 2005. PMID: 15906230 Review.
Cited by
-
Investigation of the Molecular Mechanisms by Which Endothelin-3 Stimulates Preadipocyte Growth.Front Endocrinol (Lausanne). 2021 May 21;12:661828. doi: 10.3389/fendo.2021.661828. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34093437 Free PMC article.
-
Novel immunotargets in multiple myeloma: biological relevance and therapeutic potential.Biomark Res. 2025 Jul 1;13(1):92. doi: 10.1186/s40364-025-00799-7. Biomark Res. 2025. PMID: 40597436 Free PMC article. Review.
-
Endothelin and the tumor microenvironment: a finger in every pie.Clin Sci (Lond). 2024 Jun 5;138(11):617-634. doi: 10.1042/CS20240426. Clin Sci (Lond). 2024. PMID: 38785410 Free PMC article. Review.
-
Shear Stress-Dependent Modulation of Endothelin B Receptor: The Role of Endothelial Glycocalyx Heparan Sulfate.Cells. 2025 Jul 16;14(14):1088. doi: 10.3390/cells14141088. Cells. 2025. PMID: 40710341 Free PMC article.
-
Spaceflight effects on human vascular smooth muscle cell phenotype and function.NPJ Microgravity. 2024 Mar 28;10(1):41. doi: 10.1038/s41526-024-00380-w. NPJ Microgravity. 2024. PMID: 38548798 Free PMC article.
References
-
- Rosano L, Spinella F, Bagnato A. Endothelin 1 in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2013;13(9):637–51. - PubMed
-
- Takagi Y. et al. Structural basis of G protein specificity of human endothelin receptors. A study with endothelinA/B chimeras. J Biol Chem. 1995;270(17):10072–8. - PubMed
-
- Nelson JB. et al. Identification of endothelin-1 in the pathophysiology of metastatic adenocarcinoma of the prostate. Nat Med. 1995;1(9):944–9. - PubMed
-
- Waggoner WG, Genova SL, Rash VA. Kinetic analyses demonstrate that the equilibrium assumption does not apply to [125I]endothelin-1 binding data. Life Sci. 1992;51(24):1869–76. - PubMed
LinkOut - more resources
Full Text Sources

