Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair
- PMID: 32201722
- PMCID: PMC7069697
- DOI: 10.1126/sciadv.aay4697
Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair
Erratum in
-
Erratum for the Research Article: "Global crotonylome reveals CDYL-regulated RPA1 crotonylation in homologous recombination-mediated DNA repair" by H. Yu et al.Sci Adv. 2025 Feb 7;11(6):eadv9400. doi: 10.1126/sciadv.adv9400. Epub 2025 Feb 7. Sci Adv. 2025. PMID: 39919194 Free PMC article. No abstract available.
Abstract
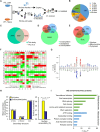
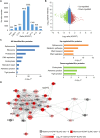
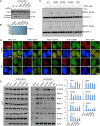
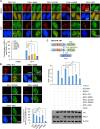
Previously, we reported that chromodomain Y-like (CDYL) acts as a crotonyl-coenzyme A hydratase and negatively regulates histone crotonylation (Kcr). However, the global CDYL-regulated crotonylome remains unclear. Here, we report a large-scale proteomics analysis for protein Kcr. We identify 14,311 Kcr sites across 3734 proteins in HeLa cells, providing by far the largest crotonylome dataset. We show that depletion of CDYL alters crotonylome landscape affecting diverse cellular pathways. Specifically, CDYL negatively regulated Kcr of RPA1, and mutation of the Kcr sites of RPA1 impaired its interaction with single-stranded DNA and/or with components of resection machinery, supporting a key role of RPA1 Kcr in homologous recombination DNA repair. Together, our study indicates that protein crotonylation has important implication in various pathophysiological processes.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures

Similar articles
-
Chromodomain Protein CDYL Acts as a Crotonyl-CoA Hydratase to Regulate Histone Crotonylation and Spermatogenesis.Mol Cell. 2017 Sep 7;67(5):853-866.e5. doi: 10.1016/j.molcel.2017.07.011. Epub 2017 Aug 10. Mol Cell. 2017. PMID: 28803779
-
Chromodomain Y-like Protein-Mediated Histone Crotonylation Regulates Stress-Induced Depressive Behaviors.Biol Psychiatry. 2019 Apr 15;85(8):635-649. doi: 10.1016/j.biopsych.2018.11.025. Epub 2018 Dec 5. Biol Psychiatry. 2019. PMID: 30665597
-
Quantitative Crotonylome Analysis Expands the Roles of p300 in the Regulation of Lysine Crotonylation Pathway.Proteomics. 2018 Aug;18(15):e1700230. doi: 10.1002/pmic.201700230. Epub 2018 Jul 11. Proteomics. 2018. PMID: 29932303 Free PMC article.
-
Lysine Crotonylation: An Emerging Player in DNA Damage Response.Biomolecules. 2022 Oct 5;12(10):1428. doi: 10.3390/biom12101428. Biomolecules. 2022. PMID: 36291637 Free PMC article. Review.
-
Emerging roles of non-histone protein crotonylation in biomedicine.Cell Biosci. 2021 May 31;11(1):101. doi: 10.1186/s13578-021-00616-2. Cell Biosci. 2021. PMID: 34059135 Free PMC article. Review.
Cited by
-
Protein lysine crotonylation in cellular processions and disease associations.Genes Dis. 2023 Aug 2;11(5):101060. doi: 10.1016/j.gendis.2023.06.029. eCollection 2024 Sep. Genes Dis. 2023. PMID: 38957707 Free PMC article. Review.
-
CapsNh-Kcr: Capsule network-based prediction of lysine crotonylation sites in human non-histone proteins.Comput Struct Biotechnol J. 2022 Dec 1;21:120-127. doi: 10.1016/j.csbj.2022.11.056. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36544479 Free PMC article.
-
A field guide to the proteomics of post-translational modifications in DNA repair.Proteomics. 2022 Aug;22(15-16):e2200064. doi: 10.1002/pmic.202200064. Epub 2022 Jun 26. Proteomics. 2022. PMID: 35695711 Free PMC article. Review.
-
Qualitative lysine crotonylome analysis in the ovarian tissue of Harmonia axyridis (Pallas).PLoS One. 2021 Oct 18;16(10):e0258371. doi: 10.1371/journal.pone.0258371. eCollection 2021. PLoS One. 2021. PMID: 34662345 Free PMC article.
-
Protein lysine crotonylation: past, present, perspective.Cell Death Dis. 2021 Jul 14;12(7):703. doi: 10.1038/s41419-021-03987-z. Cell Death Dis. 2021. PMID: 34262024 Free PMC article. Review.
References
-
- Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y., Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011). - PMC - PubMed
-
- Liu X., Wei W., Liu Y., Yang X., Wu J., Zhang Y., Zhang Q., Shi T., Du J. X., Zhao Y., Lei M., Zhou J. Q., Li J., Wong J., MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discov. 3, 17016 (2017). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

