Human Pluripotent Stem Cell-Derived Hepatocytes Show Higher Transcriptional Correlation with Adult Liver Tissue than with Fetal Liver Tissue
- PMID: 32201767
- PMCID: PMC7081255
- DOI: 10.1021/acsomega.9b03514
Human Pluripotent Stem Cell-Derived Hepatocytes Show Higher Transcriptional Correlation with Adult Liver Tissue than with Fetal Liver Tissue
Abstract
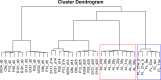
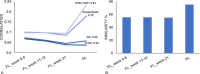
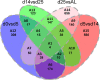
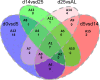
Human pluripotent stem cell-derived hepatocytes (hPSC-HEP) display many properties of mature hepatocytes, including expression of important genes of the drug metabolizing machinery, glycogen storage, and production of multiple serum proteins. To this date, hPSC-HEP do not, however, fully recapitulate the complete functionality of in vivo mature hepatocytes. In this study, we applied versatile bioinformatic algorithms, including functional annotation and pathway enrichment analyses, transcription factor binding-site enrichment, and similarity and correlation analyses, to datasets collected from different stages during hPSC-HEP differentiation and compared these to developmental stages and tissues from fetal and adult human liver. Our results demonstrate a high level of similarity between the in vitro differentiation of hPSC-HEP and in vivo hepatogenesis. Importantly, the transcriptional correlation of hPSC-HEP with adult liver (AL) tissues was higher than with fetal liver (FL) tissues (0.83 and 0.70, respectively). Functional data revealed mature features of hPSC-HEP including cytochrome P450 enzymes activities and albumin secretion. Moreover, hPSC-HEP showed expression of many genes involved in drug absorption, distribution, metabolism, and excretion. Despite the high similarities observed, we identified differences of specific pathways and regulatory players by analyzing the gene expression between hPSC-HEP and AL. These findings will aid future intervention and improvement of in vitro hepatocyte differentiation protocol in order to generate hepatocytes displaying the complete functionality of mature hepatocytes. Finally, on the transcriptional level, our results show stronger correlation and higher similarity of hPSC-HEP to AL than to FL. In addition, potential targets for further functional improvement of hPSC-HEP were also identified.
Copyright © 2020 American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): Authors Barbara Kppers-Munther, Annika Asplund ,and Christian X. Andersson are employees of Takara Bio Europe AB. Authors Tommy B. Andersson and Peter Sartipy are employees of AstraZeneca.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
