Addressing the heterogeneity in liver diseases using biological networks
- PMID: 32201876
- PMCID: PMC7986590
- DOI: 10.1093/bib/bbaa002
Addressing the heterogeneity in liver diseases using biological networks
Abstract
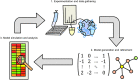
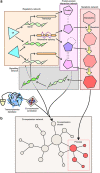
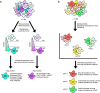
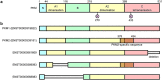
The abnormalities in human metabolism have been implicated in the progression of several complex human diseases, including certain cancers. Hence, deciphering the underlying molecular mechanisms associated with metabolic reprogramming in a disease state can greatly assist in elucidating the disease aetiology. An invaluable tool for establishing connections between global metabolic reprogramming and disease development is the genome-scale metabolic model (GEM). Here, we review recent work on the reconstruction of cell/tissue-type and cancer-specific GEMs and their use in identifying metabolic changes occurring in response to liver disease development, stratification of the heterogeneous disease population and discovery of novel drug targets and biomarkers. We also discuss how GEMs can be integrated with other biological networks for generating more comprehensive cell/tissue models. In addition, we review the various biological network analyses that have been employed for the development of efficient treatment strategies. Finally, we present three case studies in which independent studies converged on conclusions underlying liver disease.
Keywords: Computational biology; Genome-scale metabolic model; Integrated network; Liver metabolism; Omics integration; Systems biology.
© The Author(s) 2020. Published by Oxford University Press.
Figures
References
-
- Mardinoglu A, Boren J, Smith U, et al. Systems biology in hepatology: approaches and applications. Nat Rev Gastroenterol Hepatol 2018;15:365–77. - PubMed
-
- Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. - PubMed
-
- Gatto F, Volpi N, Nilsson H, et al. Glycosaminoglycan profiling in patients’ plasma and urine predicts the occurrence of metastatic clear cell renal cell carcinoma. Cell Rep 2016;15:1822–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

