Distinct effects on mRNA export factor GANP underlie neurological disease phenotypes and alter gene expression depending on intron content
- PMID: 32202298
- PMCID: PMC7297229
- DOI: 10.1093/hmg/ddaa051
Distinct effects on mRNA export factor GANP underlie neurological disease phenotypes and alter gene expression depending on intron content
Abstract
Defects in the mRNA export scaffold protein GANP, encoded by the MCM3AP gene, cause autosomal recessive early-onset peripheral neuropathy with or without intellectual disability. We extend here the phenotypic range associated with MCM3AP variants, by describing a severely hypotonic child and a sibling pair with a progressive encephalopathic syndrome. In addition, our analysis of skin fibroblasts from affected individuals from seven unrelated families indicates that disease variants result in depletion of GANP except when they alter critical residues in the Sac3 mRNA binding domain. GANP depletion was associated with more severe phenotypes compared with the Sac3 variants. Patient fibroblasts showed transcriptome alterations that suggested intron content-dependent regulation of gene expression. For example, all differentially expressed intronless genes were downregulated, including ATXN7L3B, which couples mRNA export to transcription activation by association with the TREX-2 and SAGA complexes. Our results provide insight into the molecular basis behind genotype-phenotype correlations in MCM3AP-associated disease and suggest mechanisms by which GANP defects might alter RNA metabolism.
© The Author(s) 2020. Published by Oxford University Press.
Figures



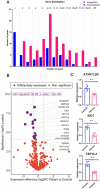

Similar articles
-
Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export.Nucleic Acids Res. 2012 May;40(10):4562-73. doi: 10.1093/nar/gks059. Epub 2012 Feb 4. Nucleic Acids Res. 2012. PMID: 22307388 Free PMC article.
-
MCM3AP in recessive Charcot-Marie-Tooth neuropathy and mild intellectual disability.Brain. 2017 Aug 1;140(8):2093-2103. doi: 10.1093/brain/awx138. Brain. 2017. PMID: 28633435
-
MCM3AP is transcribed from a promoter within an intron of the overlapping gene for GANP.J Mol Biol. 2011 Feb 25;406(3):355-61. doi: 10.1016/j.jmb.2010.12.035. Epub 2010 Dec 30. J Mol Biol. 2011. PMID: 21195085 Free PMC article.
-
Germinal Center B-Cell-Associated Nuclear Protein (GANP) Involved in RNA Metabolism for B Cell Maturation.Adv Immunol. 2016;131:135-86. doi: 10.1016/bs.ai.2016.02.003. Epub 2016 Mar 29. Adv Immunol. 2016. PMID: 27235683 Review.
-
The critical role of germinal center-associated nuclear protein in cell biology, immunohematology, and hematolymphoid oncogenesis.Exp Hematol. 2020 Oct;90:30-38. doi: 10.1016/j.exphem.2020.08.007. Epub 2020 Aug 20. Exp Hematol. 2020. PMID: 32827560 Review.
Cited by
-
Identification of biallelic mutations in MCM3AP and comprehensive literature analysis.Front Genet. 2024 Aug 20;15:1405644. doi: 10.3389/fgene.2024.1405644. eCollection 2024. Front Genet. 2024. PMID: 39228414 Free PMC article.
-
The prevalence of inherited metabolic disorders in Estonian population over 30 years: A significant increase during study period.JIMD Rep. 2022 Aug 24;63(6):604-613. doi: 10.1002/jmd2.12325. eCollection 2022 Nov. JIMD Rep. 2022. PMID: 36341167 Free PMC article.
-
Emerging Therapies and Novel Targets for TDP-43 Proteinopathy in ALS/FTD.Neurotherapeutics. 2022 Jul;19(4):1061-1084. doi: 10.1007/s13311-022-01260-5. Epub 2022 Jul 5. Neurotherapeutics. 2022. PMID: 35790708 Free PMC article. Review.
-
Genetic aspects of ataxias in a cohort of Turkish patients.Neurol Sci. 2024 Sep;45(9):4349-4365. doi: 10.1007/s10072-024-07484-x. Epub 2024 Apr 8. Neurol Sci. 2024. PMID: 38587696 Free PMC article.
References
-
- Schuurs-Hoeijmakers J.H., Vulto-van Silfhout A.T., Vissers L.E., van de V.,II, Bon B.W., Ligt J., Gilissen C., Hehir-Kwa J.Y., Neveling K., Rosario M.. et al. (2013) Identification of pathogenic gene variants in small families with intellectually disabled siblings by exome sequencing. J. Med. Genet. ,50, 802–811. - PubMed
-
- Ylikallio E., Woldegebriel R., Tumiati M., Isohanni P., Ryan M.M., Stark Z., Walsh M., Sawyer S.L., Bell K.M., Oshlack A. et al. (2017) MCM3AP in recessive charcot-marie-tooth neuropathy and mild intellectual disability. Brain, 140, 2093–2103. - PubMed
-
- Kennerson M.L., Corbett A.C., Ellis M., Perez-Siles G. and Nicholson G.A. (2018) A novel MCM3AP mutation in a Lebanese family with recessive charcot-marie-tooth neuropathy. Brain, 141, e66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

