Long non-coding RNA GRASLND enhances chondrogenesis via suppression of the interferon type II signaling pathway
- PMID: 32202492
- PMCID: PMC7202894
- DOI: 10.7554/eLife.49558
Long non-coding RNA GRASLND enhances chondrogenesis via suppression of the interferon type II signaling pathway
Abstract
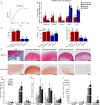
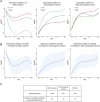
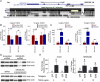
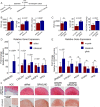
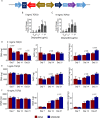
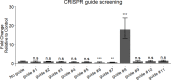
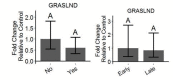
The roles of long noncoding RNAs (lncRNAs) in musculoskeletal development, disease, and regeneration remain poorly understood. Here, we identified the novel lncRNA GRASLND (originally named RNF144A-AS1) as a regulator of mesenchymal stem cell (MSC) chondrogenesis. GRASLND, a primate-specific lncRNA, is upregulated during MSC chondrogenesis and appears to act directly downstream of SOX9, but not TGF-β3. We showed that the silencing of GRASLND resulted in lower accumulation of cartilage-like extracellular matrix in a pellet assay, while GRASLND overexpression - either via transgene ectopic expression or by endogenous activation via CRISPR-dCas9-VP64 - significantly enhanced cartilage matrix production. GRASLND acts to inhibit IFN-γ by binding to EIF2AK2, and we further demonstrated that GRASLND exhibits a protective effect in engineered cartilage against interferon type II. Our results indicate an important role of GRASLND in regulating stem cell chondrogenesis, as well as its therapeutic potential in the treatment of cartilage-related diseases, such as osteoarthritis.
Keywords: RNF144A-AS1; human; mesenchymal stem cells; regenerative medicine; stem cells; tissue engineering.
© 2020, Huynh et al.
Conflict of interest statement
NH, CG, JL, RT, JB, AM, BZ, FG No competing interests declared
Figures







Comment in
-
Encouraging cartilage production.Elife. 2020 May 6;9:e57239. doi: 10.7554/eLife.57239. Elife. 2020. PMID: 32374717 Free PMC article.
References
-
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J, Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nature Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

