Learning Latent Space Representations to Predict Patient Outcomes: Model Development and Validation
- PMID: 32202503
- PMCID: PMC7136840
- DOI: 10.2196/16374
Learning Latent Space Representations to Predict Patient Outcomes: Model Development and Validation
Abstract
Background: Scalable and accurate health outcome prediction using electronic health record (EHR) data has gained much attention in research recently. Previous machine learning models mostly ignore relations between different types of clinical data (ie, laboratory components, International Classification of Diseases codes, and medications).
Objective: This study aimed to model such relations and build predictive models using the EHR data from intensive care units. We developed innovative neural network models and compared them with the widely used logistic regression model and other state-of-the-art neural network models to predict the patient's mortality using their longitudinal EHR data.
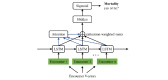
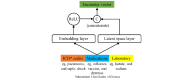
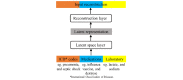
Methods: We built a set of neural network models that we collectively called as long short-term memory (LSTM) outcome prediction using comprehensive feature relations or in short, CLOUT. Our CLOUT models use a correlational neural network model to identify a latent space representation between different types of discrete clinical features during a patient's encounter and integrate the latent representation into an LSTM-based predictive model framework. In addition, we designed an ablation experiment to identify risk factors from our CLOUT models. Using physicians' input as the gold standard, we compared the risk factors identified by both CLOUT and logistic regression models.
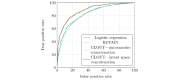
Results: Experiments on the Medical Information Mart for Intensive Care-III dataset (selected patient population: 7537) show that CLOUT (area under the receiver operating characteristic curve=0.89) has surpassed logistic regression (0.82) and other baseline NN models (<0.86). In addition, physicians' agreement with the CLOUT-derived risk factor rankings was statistically significantly higher than the agreement with the logistic regression model.
Conclusions: Our results support the applicability of CLOUT for real-world clinical use in identifying patients at high risk of mortality.
Keywords: ablation; neural networks; patient mortality; predictive modeling.
©Subendhu Rongali, Adam J Rose, David D McManus, Adarsha S Bajracharya, Alok Kapoor, Edgard Granillo, Hong Yu. Originally published in the Journal of Medical Internet Research (http://www.jmir.org), 23.03.2020.
Conflict of interest statement
Conflicts of Interest: DM has received research grant support from Apple Computer, Bristol-Myers Squibb, Boehringher-Ingelheim, Pfizer, Samsung, Philips Healthcare, Care Evolution, and Biotronik; has received consultancy fees from Bristol-Myers Squibb, Pfizer, Flexcon, and Boston Biomedical Associates; and has inventor equity in Mobile Sense Technologies, Inc, Connecticut.
Figures
Similar articles
-
A comprehensive evaluation for the prediction of mortality in intensive care units with LSTM networks: patients with cardiovascular disease.Biomed Tech (Berl). 2020 Aug 27;65(4):435-446. doi: 10.1515/bmt-2018-0206. Biomed Tech (Berl). 2020. PMID: 31846424
-
Predicting post-stroke pneumonia using deep neural network approaches.Int J Med Inform. 2019 Dec;132:103986. doi: 10.1016/j.ijmedinf.2019.103986. Epub 2019 Oct 1. Int J Med Inform. 2019. PMID: 31629312
-
Dynamic ElecTronic hEalth reCord deTection (DETECT) of individuals at risk of a first episode of psychosis: a case-control development and validation study.Lancet Digit Health. 2020 May;2(5):e229-e239. doi: 10.1016/S2589-7500(20)30024-8. Epub 2020 Mar 26. Lancet Digit Health. 2020. PMID: 33328055
-
[Prediction of intensive care unit readmission for critically ill patients based on ensemble learning].Beijing Da Xue Xue Bao Yi Xue Ban. 2021 Jun 18;53(3):566-572. doi: 10.19723/j.issn.1671-167X.2021.03.021. Beijing Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 34145862 Free PMC article. Chinese.
-
Data-driven modeling and prediction of blood glucose dynamics: Machine learning applications in type 1 diabetes.Artif Intell Med. 2019 Jul;98:109-134. doi: 10.1016/j.artmed.2019.07.007. Epub 2019 Jul 26. Artif Intell Med. 2019. PMID: 31383477 Review.
Cited by
-
Deep representation learning for clustering longitudinal survival data from electronic health records.Nat Commun. 2025 Mar 14;16(1):2534. doi: 10.1038/s41467-025-56625-z. Nat Commun. 2025. PMID: 40087274 Free PMC article.
-
Enhancing Patient Outcome Prediction Through Deep Learning With Sequential Diagnosis Codes From Structured Electronic Health Record Data: Systematic Review.J Med Internet Res. 2025 Mar 18;27:e57358. doi: 10.2196/57358. J Med Internet Res. 2025. PMID: 40100249 Free PMC article.
-
TransformEHR: transformer-based encoder-decoder generative model to enhance prediction of disease outcomes using electronic health records.Nat Commun. 2023 Nov 29;14(1):7857. doi: 10.1038/s41467-023-43715-z. Nat Commun. 2023. PMID: 38030638 Free PMC article.
-
Signals in the Cells: Multimodal and Contextualized Machine Learning Foundations for Therapeutics.bioRxiv [Preprint]. 2024 Nov 12:2024.06.12.598655. doi: 10.1101/2024.06.12.598655. bioRxiv. 2024. PMID: 38948789 Free PMC article. Preprint.
-
Prediction of Cancer Symptom Trajectory Using Longitudinal Electronic Health Record Data and Long Short-Term Memory Neural Network.JCO Clin Cancer Inform. 2024 Mar;8:e2300039. doi: 10.1200/CCI.23.00039. JCO Clin Cancer Inform. 2024. PMID: 38471054 Free PMC article.
References
-
- Choi E, Bahadori MT, Schuetz A, Stewart WF, Sun J. Doctor AI: Predicting clinical events via recurrent neural networks. JMLR Workshop Conf Proc. 2016 Aug;56:301–18. http://europepmc.org/abstract/MED/28286600 - PMC - PubMed
-
- Zhou J, Wang F, Hu J, Ye J. From Micro to Macro: Data Driven Phenotyping by Densification of Longitudinal Electronic Medical Records. Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining; KDD'14; August 24 - 27, 2014; New York, NY, USA. Association for Computing Machinery, New York, NY, United States; 2014. pp. 135–44. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

