Lipid analysis of CO2-rich subsurface aquifers suggests an autotrophy-based deep biosphere with lysolipids enriched in CPR bacteria
- PMID: 32203118
- PMCID: PMC7242380
- DOI: 10.1038/s41396-020-0624-4
Lipid analysis of CO2-rich subsurface aquifers suggests an autotrophy-based deep biosphere with lysolipids enriched in CPR bacteria
Abstract
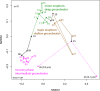
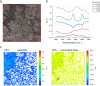
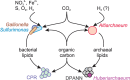
Sediment-hosted CO2-rich aquifers deep below the Colorado Plateau (USA) contain a remarkable diversity of uncultivated microorganisms, including Candidate Phyla Radiation (CPR) bacteria that are putative symbionts unable to synthesize membrane lipids. The origin of organic carbon in these ecosystems is unknown and the source of CPR membrane lipids remains elusive. We collected cells from deep groundwater brought to the surface by eruptions of Crystal Geyser, sequenced the community, and analyzed the whole community lipidome over time. Characteristic stable carbon isotopic compositions of microbial lipids suggest that bacterial and archaeal CO2 fixation ongoing in the deep subsurface provides organic carbon for the complex communities that reside there. Coupled lipidomic-metagenomic analysis indicates that CPR bacteria lack complete lipid biosynthesis pathways but still possess regular lipid membranes. These lipids may therefore originate from other community members, which also adapt to high in situ pressure by increasing fatty acid unsaturation. An unusually high abundance of lysolipids attributed to CPR bacteria may represent an adaptation to membrane curvature stress induced by their small cell sizes. Our findings provide new insights into the carbon cycle in the deep subsurface and suggest the redistribution of lipids into putative symbionts within this community.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface.Nat Microbiol. 2018 Mar;3(3):328-336. doi: 10.1038/s41564-017-0098-y. Epub 2018 Jan 29. Nat Microbiol. 2018. PMID: 29379208 Free PMC article.
-
Metagenomic analysis of a high carbon dioxide subsurface microbial community populated by chemolithoautotrophs and bacteria and archaea from candidate phyla.Environ Microbiol. 2016 Jun;18(6):1686-703. doi: 10.1111/1462-2920.12817. Epub 2015 Apr 8. Environ Microbiol. 2016. PMID: 25727367
-
Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities.Microbiome. 2017 Sep 2;5(1):112. doi: 10.1186/s40168-017-0331-1. Microbiome. 2017. PMID: 28865481 Free PMC article.
-
Microbial ecology of the deep terrestrial subsurface.ISME J. 2024 Jan 8;18(1):wrae091. doi: 10.1093/ismejo/wrae091. ISME J. 2024. PMID: 38780093 Free PMC article. Review.
-
[Thermophilic prokaryotes from deep subterranean habitats].Mikrobiologiia. 2014 May-Jun;83(3):255-70. Mikrobiologiia. 2014. PMID: 25844436 Review. Russian.
Cited by
-
Ultra-small bacteria and archaea exhibit genetic flexibility towards groundwater oxygen content, and adaptations for attached or planktonic lifestyles.ISME Commun. 2023 Feb 17;3(1):13. doi: 10.1038/s43705-023-00223-x. ISME Commun. 2023. PMID: 36808147 Free PMC article.
-
Leave no stone unturned: individually adapted xerotolerant Thaumarchaeota sheltered below the boulders of the Atacama Desert hyperarid core.Microbiome. 2021 Nov 26;9(1):234. doi: 10.1186/s40168-021-01177-9. Microbiome. 2021. PMID: 34836555 Free PMC article.
-
Cooperation and Competition Were Primary Driving Forces for Biological Evolution.Microb Physiol. 2025;35(1):13-29. doi: 10.1159/000544890. Epub 2025 Feb 25. Microb Physiol. 2025. PMID: 39999802 Review.
-
Reductive evolution and unique predatory mode in the CPR bacterium Vampirococcus lugosii.Nat Commun. 2021 Apr 28;12(1):2454. doi: 10.1038/s41467-021-22762-4. Nat Commun. 2021. PMID: 33911080 Free PMC article.
-
Production of structurally diverse sphingolipids by anaerobic marine bacteria in the euxinic Black Sea water column.ISME J. 2024 Jan 8;18(1):wrae153. doi: 10.1093/ismejo/wrae153. ISME J. 2024. PMID: 39113610 Free PMC article.
References
-
- Orsi WD. Ecology and evolution of seafloor and subseafloor microbial communities. Nat Rev Microbiol. 2018;16:671–83. - PubMed
-
- Magnabosco C, Lin L-H, Dong H, Bomberg M, Ghiorse W, Stan-Lotter H, et al. The biomass and biodiversity of the continental subsurface. Nat Geosci. 2018;11:707.

