Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability
- PMID: 32203227
- PMCID: PMC7222126
- DOI: 10.1038/s41436-020-0771-z
Systematic evidence-based review: outcomes from exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability
Abstract
Purpose: Exome and genome sequencing (ES/GS) are performed frequently in patients with congenital anomalies, developmental delay, or intellectual disability (CA/DD/ID), but the impact of results from ES/GS on clinical management and patient outcomes is not well characterized. A systematic evidence review (SER) can support future evidence-based guideline development for use of ES/GS in this patient population.
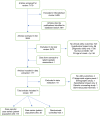
Methods: We undertook an SER to identify primary literature from January 2007 to March 2019 describing health, clinical, reproductive, and psychosocial outcomes resulting from ES/GS in patients with CA/DD/ID. A narrative synthesis of results was performed.
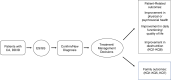
Results: We retrieved 2654 publications for full-text review from 7178 articles. Only 167 articles met our inclusion criteria, and these were primarily case reports or small case series of fewer than 20 patients. The most frequently reported outcomes from ES/GS were changes to clinical management or reproductive decision-making. Two studies reported on the reduction of mortality or morbidity or impact on quality of life following ES/GS.
Conclusion: There is evidence that ES/GS for patients with CA/DD/ID informs clinical and reproductive decision-making, which could lead to improved outcomes for patients and their family members. Further research is needed to generate evidence regarding health outcomes to inform robust guidelines regarding ES/GS in the care of patients with CA/DD/ID.
Keywords: clinical genetics; congenital anomalies; exome sequencing; intellectual disability; systematic evidence review.
Conflict of interest statement
S.E.H. received research support from Alexion Pharmaceuticals to serve as a site investigator for a prospective registry for hypophosphatasia. D.T.M. received honoraria from Prevention Genetics and Takeda Pharmaceuticals. M.C.S. receives salary (no equity) as a lab director at Washington University School of Medicine, received salary (no equity) as a lab director at HudsonAlpha Institute of Biotechnology (performs clinical whole genome sequencing), and received consulting fees from PierianDx. J.S. received salary from the Partners Healthcare Laboratory for Molecular Medicine and from the Brigham and Women’s Hospital Cytogenomics Laboratory. The other authors declare no conflicts of interest.
Figures
References
-
- Hochstenbach R, van Binsbergen E, Engelen J, et al. Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet. 2009;52:161–169. - PubMed
-
- Fan LL, Huang H, Jin JY, et al. Whole exome sequencing identifies a novel mutation (c.333+2T>C) of TNNI3K in a Chinese family with dilated cardiomyopathy and cardiac conduction disease. Gene. 2018;648:63–67. - PubMed
-
- Cheng SSW, Chan KYK, Leung KKP, et al. Experience of chromosomal microarray applied in prenatal and postnatal settings in Hong Kong. Am J Med Genet C Semin Med Genet. 2019;181:196–207. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

