Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity
- PMID: 32203410
- PMCID: PMC7190418
- DOI: 10.1038/s41564-020-0691-3
Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity
Abstract
CRISPR-Cas systems are adaptive immune systems that protect bacteria from bacteriophage (phage) infection1. To provide immunity, RNA-guided protein surveillance complexes recognize foreign nucleic acids, triggering their destruction by Cas nucleases2. While the essential requirements for immune activity are well understood, the physiological cues that regulate CRISPR-Cas expression are not. Here, a forward genetic screen identifies a two-component system (KinB-AlgB), previously characterized in the regulation of Pseudomonas aeruginosa alginate biosynthesis3,4, as a regulator of the expression and activity of the P. aeruginosa Type I-F CRISPR-Cas system. Downstream of KinB-AlgB, activators of alginate production AlgU (a σE orthologue) and AlgR repress CRISPR-Cas activity during planktonic and surface-associated growth5. AmrZ, another alginate regulator6, is triggered to repress CRISPR-Cas immunity upon surface association. Pseudomonas phages and plasmids have taken advantage of this regulatory scheme and carry hijacked homologs of AmrZ that repress CRISPR-Cas expression and activity. This suggests that while CRISPR-Cas regulation may be important to limit self-toxicity, endogenous repressive pathways represent a vulnerability for parasite manipulation.
Conflict of interest statement
Competing Interests
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences.
Figures


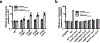
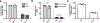


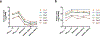


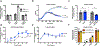
Similar articles
-
Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa.J Bacteriol. 1998 Feb;180(4):956-68. doi: 10.1128/JB.180.4.956-968.1998. J Bacteriol. 1998. PMID: 9473053 Free PMC article.
-
Clustered Regularly Interspaced Short Palindromic Repeat-Dependent, Biofilm-Specific Death of Pseudomonas aeruginosa Mediated by Increased Expression of Phage-Related Genes.mBio. 2015 May 12;6(3):e00129-15. doi: 10.1128/mBio.00129-15. mBio. 2015. PMID: 25968642 Free PMC article.
-
Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT.J Bacteriol. 1994 Oct;176(19):6007-14. doi: 10.1128/jb.176.19.6007-6014.1994. J Bacteriol. 1994. PMID: 7928961 Free PMC article.
-
The Biology of CRISPR-Cas: Backward and Forward.Cell. 2018 Mar 8;172(6):1239-1259. doi: 10.1016/j.cell.2017.11.032. Cell. 2018. PMID: 29522745 Review.
-
Meet the Anti-CRISPRs: Widespread Protein Inhibitors of CRISPR-Cas Systems.CRISPR J. 2019 Feb;2(1):23-30. doi: 10.1089/crispr.2018.0052. CRISPR J. 2019. PMID: 31021234 Review.
Cited by
-
Biomimetic natural biomaterials for tissue engineering and regenerative medicine: new biosynthesis methods, recent advances, and emerging applications.Mil Med Res. 2023 Mar 28;10(1):16. doi: 10.1186/s40779-023-00448-w. Mil Med Res. 2023. PMID: 36978167 Free PMC article. Review.
-
Phage Therapy: A Different Approach to Fight Bacterial Infections.Biologics. 2022 Oct 6;16:173-186. doi: 10.2147/BTT.S381237. eCollection 2022. Biologics. 2022. PMID: 36225325 Free PMC article. Review.
-
Phages against Pathogenic Bacterial Biofilms and Biofilm-Based Infections: A Review.Pharmaceutics. 2022 Feb 16;14(2):427. doi: 10.3390/pharmaceutics14020427. Pharmaceutics. 2022. PMID: 35214158 Free PMC article. Review.
-
Anti-CRISPRs go viral: The infection biology of CRISPR-Cas inhibitors.Cell Host Microbe. 2021 May 12;29(5):704-714. doi: 10.1016/j.chom.2020.12.007. Epub 2021 Jan 13. Cell Host Microbe. 2021. PMID: 33444542 Free PMC article. Review.
-
Oxidative stress elicited by phage infection induces Staphylococcal type III-A CRISPR-Cas system.Nucleic Acids Res. 2025 Jun 20;53(12):gkaf541. doi: 10.1093/nar/gkaf541. Nucleic Acids Res. 2025. PMID: 40548936 Free PMC article.
References
-
- Barrangou R et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). - PubMed

