Genetic Transformation of Tribonema minus, a Eukaryotic Filamentous Oleaginous Yellow-Green Alga
- PMID: 32204356
- PMCID: PMC7139823
- DOI: 10.3390/ijms21062106
Genetic Transformation of Tribonema minus, a Eukaryotic Filamentous Oleaginous Yellow-Green Alga
Abstract
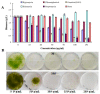
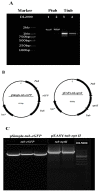
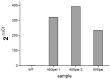
Eukaryotic filamentous yellow-green algae from the Tribonema genus are considered to be excellent candidates for biofuels and value-added products, owing to their ability to grow under autotrophic, mixotrophic, and heterotrophic conditions and synthesize large amounts of fatty acids, especially unsaturated fatty acids. To elucidate the molecular mechanism of fatty acids and/or establish the organism as a model strain, the development of genetic methods is important. Towards this goal, here, we constructed a genetic transformation method to introduce exogenous genes for the first time into the eukaryotic filamentous alga Tribonema minus via particle bombardment. In this study, we constructed pSimple-tub-eGFP and pEASY-tub-nptⅡ plasmids in which the green fluorescence protein (eGFP) gene and the neomycin phosphotransferase Ⅱ-encoding G418-resistant gene (nptⅡ) were flanked by the T. minus-derived tubulin gene (tub) promoter and terminator, respectively. The two plasmids were introduced into T. minus cells through particle-gun bombardment under various test conditions. By combining agar and liquid selecting methods to exclude the pseudotransformants under long-term antibiotic treatment, plasmids pSimple-tub-eGFP and pEASY-tub- nptⅡ were successfully transformed into the genome of T. minus, which was verified using green fluorescence detection and the polymerase chain reaction, respectively. These results suggest new possibilities for efficient genetic engineering of T. minus for future genetic improvement.
Keywords: Tribonema minus; green fluorescence protein; particle-gun bombardment; transformation; tubulin gene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Irina A.G., John L.H. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 2006;45:160–186. - PubMed
-
- William P.L.I.B., Laurens L.M.L. Microalgae as biodiesel and biomass feedstocks: Review and analysis of the biochemistry, energetics and economics. Energy Environ. Sci. 2010;3:554–590. doi: 10.1039/b924978h. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

