A Quinolinone Compound Inhibiting the Oligomerization of Nucleoprotein of Influenza A Virus Prevents the Selection of Escape Mutants
- PMID: 32204549
- PMCID: PMC7150793
- DOI: 10.3390/v12030337
A Quinolinone Compound Inhibiting the Oligomerization of Nucleoprotein of Influenza A Virus Prevents the Selection of Escape Mutants
Abstract
The emergence of resistance to currently available anti-influenza drugs has heightened the need for antivirals with novel mechanisms of action. The influenza A virus (IAV) nucleoprotein (NP) is highly conserved and essential for the formation of viral ribonucleoprotein (vRNP), which serves as the template for replication and transcription. Recently, using in silico screening, we identified an antiviral compound designated NUD-1 (a 4-hydroxyquinolinone derivative) as a potential inhibitor of NP. In this study, we further analyzed the interaction between NUD-1 and NP and found that the compound interferes with the oligomerization of NP, which is required for vRNP formation, leading to the suppression of viral transcription, protein synthesis, and nuclear export of NP. We further assessed the selection of resistant variants by serially passaging a clinical isolate of the 2009 H1N1 pandemic influenza virus in the presence of NUD-1 or oseltamivir. NUD-1 did not select for resistant variants after nine passages, whereas oseltamivir selected for resistant variants after five passages. Our data demonstrate that NUD-1 interferes with the oligomerization of NP and less likely induces drug-resistant variants than oseltamivir; hence, it is a potential lead compound for the development of novel anti-influenza drugs.
Keywords: 4-hydroxyquinolinone; antiviral; nucleoprotein; oligomerization; resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



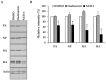
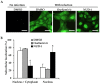
References
-
- Hurt A.C., Ernest J., Deng Y.-M., Iannello P., Besselaar T.G., Birch C., Buchy P., Chittaganpitch M., Chiu S.C., Dwyer D., et al. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antivir. Res. 2009;83:90–93. doi: 10.1016/j.antiviral.2009.03.003. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

