Fis Contributes to Resistance of Pseudomonas aeruginosa to Ciprofloxacin by Regulating Pyocin Synthesis
- PMID: 32205461
- PMCID: PMC7221260
- DOI: 10.1128/JB.00064-20
Fis Contributes to Resistance of Pseudomonas aeruginosa to Ciprofloxacin by Regulating Pyocin Synthesis
Abstract
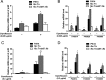
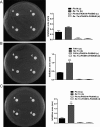
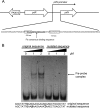
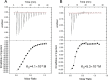
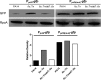
Factor for inversion stimulation (Fis) is a versatile DNA binding protein that plays an important role in coordinating bacterial global gene expression in response to growth phases and environmental stresses. Previously, we demonstrated that Fis regulates the type III secretion system (T3SS) in Pseudomonas aeruginosa In this study, we explored the role of Fis in the antibiotic resistance of P. aeruginosa and found that mutation of the fis gene increases the bacterial susceptibility to ciprofloxacin. We further demonstrated that genes related to pyocin biosynthesis are upregulated in the fis mutant. The pyocins are produced in response to genotoxic agents, including ciprofloxacin, and the release of pyocins results in lysis of the producer cell. Thus, pyocin biosynthesis genes sensitize P. aeruginosa to ciprofloxacin. We found that PrtN, the positive regulator of the pyocin biosynthesis genes, is upregulated in the fis mutant. Genetic experiments and electrophoretic mobility shift assays revealed that Fis directly binds to the promoter region of prtN and represses its expression. Therefore, our results revealed novel Fis-mediated regulation on pyocin production and bacterial resistance to ciprofloxacin in P. aeruginosaIMPORTANCEPseudomonas aeruginosa is an important opportunistic pathogenic bacterium that causes various acute and chronic infections in human, especially in patients with compromised immunity, cystic fibrosis (CF), and/or severe burn wounds. About 60% of cystic fibrosis patients have a chronic respiratory infection caused by P. aeruginosa The bacterium is intrinsically highly resistant to antibiotics, which greatly increases difficulties in clinical treatment. Therefore, it is critical to understand the mechanisms and the regulatory pathways that are involved in antibiotic resistance. In this study, we elucidated a novel regulatory pathway that controls the bacterial resistance to fluoroquinolone antibiotics, which enhances our understanding of how P. aeruginosa responds to ciprofloxacin.
Keywords: Fis; Pseudomonas aeruginosa; pyocin; resistance.
Copyright © 2020 American Society for Microbiology.
Figures
References
-
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

