Circular RNA-protein interactions: functions, mechanisms, and identification
- PMID: 32206104
- PMCID: PMC7069073
- DOI: 10.7150/thno.42174
Circular RNA-protein interactions: functions, mechanisms, and identification
Abstract
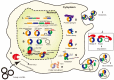
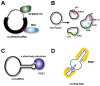
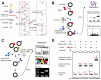
Circular RNAs (circRNAs) are covalently closed, endogenous RNAs with no 5' end caps or 3' poly(A) tails. These RNAs are expressed in tissue-specific, cell-specific, and developmental stage-specific patterns. The biogenesis of circRNAs is now known to be regulated by multiple specific factors; however, circRNAs were previously thought to be insignificant byproducts of splicing errors. Recent studies have demonstrated their activity as microRNA (miRNA) sponges as well as protein sponges, decoys, scaffolds, and recruiters, and some circRNAs even act as translation templates in multiple pathophysiological processes. CircRNAs bind and sequester specific proteins to appropriate subcellular positions, and they participate in modulating certain protein-protein and protein-RNA interactions. Conversely, several proteins play an indispensable role in the life cycle of circRNAs from biogenesis to degradation. However, the exact mechanisms of these interactions between proteins and circRNAs remain unknown. Here, we review the current knowledge regarding circRNA-protein interactions and the methods used to identify and characterize these interactions. We also summarize new insights into the potential mechanisms underlying these interactions.
Keywords: RNA binding proteins (RBPs); biogenesis; circular RNAs (circRNAs); degradation; translation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

