Recombinase polymerase amplification assay for the detection of piper yellow mottle virus infecting black pepper
- PMID: 32206697
- PMCID: PMC7085498
- DOI: 10.1007/s13337-019-00566-x
Recombinase polymerase amplification assay for the detection of piper yellow mottle virus infecting black pepper
Abstract
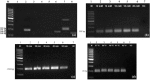
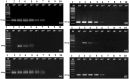
Recombinase polymerase amplification (RPA) is a quick, specific, sensitive molecular tool carried out at a constant temperature for pathogen detection. In the present study, RPA and reverse transcription (RT) RPA assays were optimized for the detection of piper yellow mottle virus (PYMoV) infecting black pepper. Out of the eight primer pairs targeted to amplify open reading frames (ORFs) 2 and 3 of the virus, the primer pair targeted to ORF2 gave specific amplification only with DNA isolated from infected plant but not with healthy plant. A magnesium acetate concentration of 18 mM, 40 min of incubation time and a temperature of 37-42 °C was found optimum for detection of the virus in RPA assay. Comparison of sensitivity of detection revealed that RPA could detect the virus up to 10-5 dilution of the total DNA while PCR could detect the virus up to 10-4 dilution indicating that RPA is 10 times more sensitive than PCR. RPA was further simplified using crude extract as template which could detect the virus up to 10-3 dilution. RT-RPA was optimized for the detection of PYMoV using total RNA isolated from infected plants as the template. Both RT-RPA and RPA assays were validated using field samples of black pepper representing different varieties and geographical regions by using CTAB isolated DNA, crude DNA extract and cDNA. Our study showed that RPA and RT-RPA can be successfully adopted as a substitute to PCR for detection of PYMoV infecting black pepper.
Keywords: Diagnosis; Isothermal amplification; Polymerase chain reaction; Reverse transcription-RPA; Sensitivity.
© Indian Virological Society 2020.
Conflict of interest statement
Conflict of interestBoth authors declare that they have no conflict of interest.
Figures


References
-
- Babu B, Washburn BK, Ertek TS, Miller SH, Riddle CB, Knox GW, Ochoa-Corona FM, Olson J, Katırcıoğlu YZ, Paret ML. A field based detection method for Rose rosette virus using isothermal probe based reverse transcription-recombinase polymerase amplification assay. J Virol Methods. 2017;247:81–90. doi: 10.1016/j.jviromet.2017.05.019. - DOI - PubMed
-
- Bhat AI, Siljo A. Detection of viruses infecting black pepper by SYBR Green based real-time PCR assay. Plant Pathol J. 2014;96:105–109.
-
- Bhat AI, Siljo A, Jiby MV, Thankamani CK, Mathew PA. Polymerase chain reaction (PCR) based indexing for screening black pepper (Piper nigrum L.) plants against Piper yellow mottle virus. J Spices Aromat Crop. 2009;18:28–32.
LinkOut - more resources
Full Text Sources

