Rhodobacter capsulatus AnfA is essential for production of Fe-nitrogenase proteins but dispensable for cofactor biosynthesis and electron supply
- PMID: 32207246
- PMCID: PMC7294313
- DOI: 10.1002/mbo3.1033
Rhodobacter capsulatus AnfA is essential for production of Fe-nitrogenase proteins but dispensable for cofactor biosynthesis and electron supply
Abstract
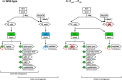
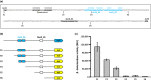
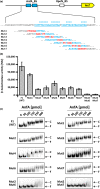
The photosynthetic α-proteobacterium Rhodobacter capsulatus reduces and thereby fixes atmospheric dinitrogen (N2 ) by a molybdenum (Mo)-nitrogenase and an iron-only (Fe)-nitrogenase. Differential expression of the structural genes of Mo-nitrogenase (nifHDK) and Fe-nitrogenase (anfHDGK) is strictly controlled and activated by NifA and AnfA, respectively. In contrast to NifA-binding sites, AnfA-binding sites are poorly defined. Here, we identified two highly similar AnfA-binding sites in the R. capsulatus anfH promoter by studying the effects of promoter mutations on in vivo anfH expression and in vitro promoter binding by AnfA. Comparison of the experimentally determined R. capsulatus AnfA-binding sites and presumed AnfA-binding sites from other α-proteobacteria revealed a consensus sequence of dyad symmetry, TAC-N6 -GTA, suggesting that AnfA proteins bind their target promoters as dimers. Chromosomal replacement of the anfH promoter by the nifH promoter restored anfHDGK expression and Fe-nitrogenase activity in an R. capsulatus strain lacking AnfA suggesting that AnfA is required for AnfHDGK production, but dispensable for biosynthesis of the iron-only cofactor and electron delivery to Fe-nitrogenase, pathways activated by NifA. These observations strengthen our model, in which the Fe-nitrogenase system in R. capsulatus is largely integrated into the Mo-nitrogenase system.
Keywords: Rhodobacter; AnfA; Fe-nitrogenase; Mo-nitrogenase; NifA.
© 2020 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Austin, S. , & Lambert, J. (1994). Purification and in vitro activity of a truncated form of ANFA. Transcriptional activator protein of alternative nitrogenase from Azotobacter vinelandii . Journal of Biological Chemistry, 269, 18141–18148. - PubMed
-
- Barrios, H. , Grande, R. , Olvera, L. , & Morett, E. (1998). In vivo genomic footprinting analysis reveals that the complex Bradyrhizobium japonicum fixRnifA promoter region is differently occupied by two distinct RNA polymerase holoenzymes. Proceedings of the National Academy of Sciences of the United States of America, 95, 1014–1019. - PMC - PubMed
-
- Buck, M. , Miller, S. , Drummond, M. , & Dixon, R. (1986). Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature, 320, 374–378. 10.1038/320374a0 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

