CENP-F stabilizes kinetochore-microtubule attachments and limits dynein stripping of corona cargoes
- PMID: 32207772
- PMCID: PMC7199848
- DOI: 10.1083/jcb.201905018
CENP-F stabilizes kinetochore-microtubule attachments and limits dynein stripping of corona cargoes
Abstract
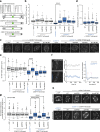
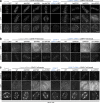
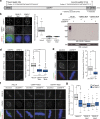
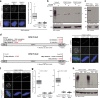
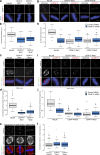
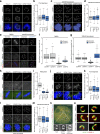
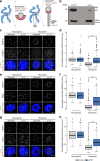
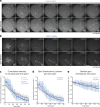
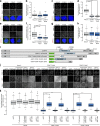
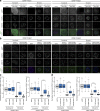
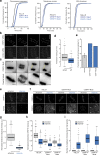
Accurate chromosome segregation demands efficient capture of microtubules by kinetochores and their conversion to stable bioriented attachments that can congress and then segregate chromosomes. An early event is the shedding of the outermost fibrous corona layer of the kinetochore following microtubule attachment. Centromere protein F (CENP-F) is part of the corona, contains two microtubule-binding domains, and physically associates with dynein motor regulators. Here, we have combined CRISPR gene editing and engineered separation-of-function mutants to define how CENP-F contributes to kinetochore function. We show that the two microtubule-binding domains make distinct contributions to attachment stability and force transduction but are dispensable for chromosome congression. We further identify a specialized domain that functions to limit the dynein-mediated stripping of corona cargoes through a direct interaction with Nde1. This antagonistic activity is crucial for maintaining the required corona composition and ensuring efficient kinetochore biorientation.
© 2020 Auckland et al.
Figures




References
-
- Allan L.A., Reis M., Liu Y., Huis in ’t Veld P., Kops G.J.P.L., Musacchio A., and Saurin A.T.. 2019. Cyclin B1 scaffolds MAD1 at the corona to activate the spindle assembly checkpoint. bioRxiv. doi:10.1101/726224 (Preprint posted August 5, 2019)

