Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome
- PMID: 32207811
- PMCID: PMC7971136
- DOI: 10.1084/jem.20191804
Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome
Erratum in
-
Correction: Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome.J Exp Med. 2020 Jun 1;217(7):e2019180405272020c. doi: 10.1084/jem.2019180405272020c. J Exp Med. 2020. PMID: 32516385 Free PMC article. No abstract available.
Abstract
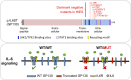
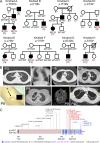
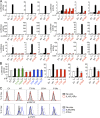
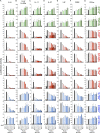
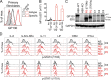
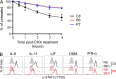
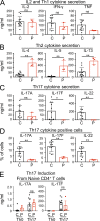
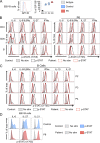
Autosomal dominant hyper-IgE syndrome (AD-HIES) is typically caused by dominant-negative (DN) STAT3 mutations. Patients suffer from cold staphylococcal lesions and mucocutaneous candidiasis, severe allergy, and skeletal abnormalities. We report 12 patients from 8 unrelated kindreds with AD-HIES due to DN IL6ST mutations. We identified seven different truncating mutations, one of which was recurrent. The mutant alleles encode GP130 receptors bearing the transmembrane domain but lacking both the recycling motif and all four STAT3-recruiting tyrosine residues. Upon overexpression, the mutant proteins accumulate at the cell surface and are loss of function and DN for cellular responses to IL-6, IL-11, LIF, and OSM. Moreover, the patients' heterozygous leukocytes and fibroblasts respond poorly to IL-6 and IL-11. Consistently, patients with STAT3 and IL6ST mutations display infectious and allergic manifestations of IL-6R deficiency, and some of the skeletal abnormalities of IL-11R deficiency. DN STAT3 and IL6ST mutations thus appear to underlie clinical phenocopies through impairment of the IL-6 and IL-11 response pathways.
© 2020 Béziat et al.
Conflict of interest statement
Disclosures: Dr. Chen reported grants from Bristol-Myers Squibb during the conduct of the study. Dr. Rosenfeld reported personal fees from Baylor Genetics Laboratories outside the submitted work. Dr. Milner reported a patent to use STAT3 inhibition to prevent anaphylaxis pending. Dr. Couderc reported non-financial support from Astra Zeneca, personal fees from Boehringer Ingelheim, personal fees from Novartis, and grants from LVL outside the submitted work. Dr. Catherinot reported financial support for travel and registration expenses related to international medical meetings (LVL Medical, CSL Behring). Dr. Uhlig reported grants from Celgene during the conduct of the study and grants from UCB and Eli Lilly outside the submitted work. Dr. Haerynck reported, "Centre for Primary Immune deficiency is recognized as a Jeffrey Modell Foundation diagnostic and research center and supported by the Jeffrey Modell Foundation; the University Hospital Ghent Spearhead Initiative for Immunology Research (until 7/2019); the Grand Challenges Program of VIB (this VIB Program received support from the Flemish Government under the Management Agreement 2017-2021; VR 2016 2312 Doc.1521/4); Simon Tavernier is a postdoctoral fellow at PID research lab with the Fund for Scientific Research Flanders (FWO, 12W9119N); I am funded by a university research grant (BOF-University Ghent)." No other disclosures were reported.
Figures







References
-
- Arita, K., South A.P., Hans-Filho G., Sakuma T.H., Lai-Cheong J., Clements S., Odashiro M., Odashiro D.N., Hans-Neto G., Hans N.R., et al. . 2008. Oncostatin M receptor-β mutations underlie familial primary localized cutaneous amyloidosis. Am. J. Hum. Genet. 82:73–80. 10.1016/j.ajhg.2007.09.002 - DOI - PMC - PubMed
-
- Avery, D.T., Deenick E.K., Ma C.S., Suryani S., Simpson N., Chew G.Y., Chan T.D., Palendira U., Bustamante J., Boisson-Dupuis S., et al. . 2010. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 207:155–171. 10.1084/jem.20091706 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

