A bacteriophage mimic of the bacterial nucleoid-associated protein Fis
- PMID: 32207815
- PMCID: PMC7166090
- DOI: 10.1042/BCJ20200146
A bacteriophage mimic of the bacterial nucleoid-associated protein Fis
Abstract
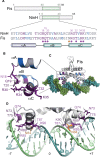
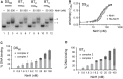
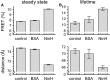
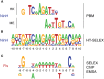
We report the identification and characterization of a bacteriophage λ-encoded protein, NinH. Sequence homology suggests similarity between NinH and Fis, a bacterial nucleoid-associated protein (NAP) involved in numerous DNA topology manipulations, including chromosome condensation, transcriptional regulation and phage site-specific recombination. We find that NinH functions as a homodimer and is able to bind and bend double-stranded DNA in vitro. Furthermore, NinH shows a preference for a 15 bp signature sequence related to the degenerate consensus favored by Fis. Structural studies reinforced the proposed similarity to Fis and supported the identification of residues involved in DNA binding which were demonstrated experimentally. Overexpression of NinH proved toxic and this correlated with its capacity to associate with DNA. NinH is the first example of a phage-encoded Fis-like NAP that likely influences phage excision-integration reactions or bacterial gene expression.
Keywords: DNA bending; DNA binding; Fis; NinH; bacteriophage λ; nucleoid-associated proteins.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

