An enzymatic on/off switch-mediated assay for KRAS hotspot point mutation detection of circulating tumor DNA
- PMID: 32207862
- PMCID: PMC7439329
- DOI: 10.1002/jcla.23305
An enzymatic on/off switch-mediated assay for KRAS hotspot point mutation detection of circulating tumor DNA
Abstract
Background: To detect the mutations of KRAS gene in colorectal cancer patients and other cancer patients, it is of value to develop non-invasive, sensitive, specific, easy, and low-cost assays.
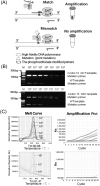
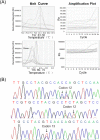
Methods: Templates harboring hotspot mutations of the KRAS gene were constructed, and primers were designed for evaluation of the specificity, and sensitivity of detection system consisted of exonuclease polymerase-mediated on/off switch; then, gel electrophoresis and real-time PCR were performed for verification. The assay was verified by testing the DNA pool of normal controls and circulating DNA (ctDNA) samples from 14 tumor patients, as compared to Sanger sequencing.
Results: A specific and sensitive assay consisted of exonuclease polymerase-mediated on/off switch, and multiplex real-time PCR method has been established. This assay could detect <100 copies of KRAS mutation in more than 10 million copies of wild-type KRAS gene fragments. This assay was applied to test KRAS gene mutations in three cases of fourteen ctDNA samples, and the results were consistent with Sanger sequencing. However, this PCR-based assay was more sensitive and easier to be interpreted.
Conclusion: This assay can detect the presence of KRAS hotspot mutations in clinical circulating tumor DNA samples. The assay has a potential to be used in early diagnosis of colorectal cancer as well as other types of cancer.
Keywords: KRAS mutations; ctDNA; enzymatic on/off switch; multiplex PCR; mutation detection.
© 2020 The Authors. Journal of Clinical Laboratory Analysis published by Wiley Periodicals LLC.
Conflict of interest statement
No competing financial interests exist.
Figures

Similar articles
-
A novel KRAS exon 2 drop-off digital PCR assay for mutation detection in cell-free DNA of cancer patients.Diagn Pathol. 2025 May 24;20(1):62. doi: 10.1186/s13000-025-01637-y. Diagn Pathol. 2025. PMID: 40413426 Free PMC article.
-
Multiplex detection of ctDNA mutations in plasma of colorectal cancer patients by PCR/SERS assay.Nanotheranostics. 2020 Aug 25;4(4):224-232. doi: 10.7150/ntno.48905. eCollection 2020. Nanotheranostics. 2020. PMID: 32923312 Free PMC article.
-
Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing.Sci Rep. 2020 May 15;10(1):8122. doi: 10.1038/s41598-020-64822-7. Sci Rep. 2020. PMID: 32415199 Free PMC article.
-
The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: A systematic review and meta-analysis.PLoS One. 2021 Mar 26;16(3):e0248775. doi: 10.1371/journal.pone.0248775. eCollection 2021. PLoS One. 2021. PMID: 33770081 Free PMC article.
-
Circulating tumor DNA is effective for detection of KRAS mutation in colorectal cancer: a meta-analysis.Int J Biol Markers. 2017 Oct 31;32(4):e421-e427. doi: 10.5301/ijbm.5000295. Int J Biol Markers. 2017. PMID: 28885658
Cited by
-
KMT2D promotes proliferation of gastric cancer cells: evidence from ctDNA sequencing.J Clin Lab Anal. 2021 Apr;35(4):e23721. doi: 10.1002/jcla.23721. Epub 2021 Apr 1. J Clin Lab Anal. 2021. PMID: 33793001 Free PMC article.
References
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789‐799. - PubMed
-
- Adjei AA. Blocking oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2002;94(13):1031‐1032. - PubMed
-
- Neumann J, Zeindl‐Eberhart E, Kirchner T, et al. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205(12):858‐862. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

