A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat
- PMID: 32208137
- PMCID: PMC7093151
- DOI: 10.7554/eLife.55646
A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat
Abstract
Understanding the function of genes within staple crops will accelerate crop improvement by allowing targeted breeding approaches. Despite their importance, a lack of genomic information and resources has hindered the functional characterisation of genes in major crops. The recent release of high-quality reference sequences for these crops underpins a suite of genetic and genomic resources that support basic research and breeding. For wheat, these include gene model annotations, expression atlases and gene networks that provide information about putative function. Sequenced mutant populations, improved transformation protocols and structured natural populations provide rapid methods to study gene function directly. We highlight a case study exemplifying how to integrate these resources. This review provides a helpful guide for plant scientists, especially those expanding into crop research, to capitalise on the discoveries made in Arabidopsis and other plants. This will accelerate the improvement of crops of vital importance for food and nutrition security.
Keywords: crop genetics; genomics; plant biology; polyploidy; wheat.
© 2020, Adamski et al.
Conflict of interest statement
NA, PB, JB, SH, CM, AB, WB, LC, JC, BC, BF, SG, WH, KH, SH, LH, KK, JK, MM, GN, CP, RR, CS, BT, LW, BW, CU No competing interests declared
Figures

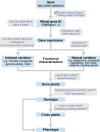
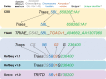



References
-
- Alaux M, Rogers J, Letellier T, Flores R, Alfama F, Pommier C, Mohellibi N, Durand S, Kimmel E, Michotey C, Guerche C, Loaec M, Lainé M, Steinbach D, Choulet F, Rimbert H, Leroy P, Guilhot N, Salse J, Feuillet C, Paux E, Eversole K, Adam-Blondon AF, Quesneville H, International Wheat Genome Sequencing Consortium Linking the international wheat genome sequencing consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biology. 2018;19:111. doi: 10.1186/s13059-018-1491-4. - DOI - PMC - PubMed
-
- Allen AM, Barker GL, Berry ST, Coghill JA, Gwilliam R, Kirby S, Robinson P, Brenchley RC, D'Amore R, McKenzie N, Waite D, Hall A, Bevan M, Hall N, Edwards KJ. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.) Plant Biotechnology Journal. 2011;9:1086–1099. doi: 10.1111/j.1467-7652.2011.00628.x. - DOI - PubMed
-
- Allen AM, Winfield MO, Burridge AJ, Downie RC, Benbow HR, Barker GL, Wilkinson PA, Coghill J, Waterfall C, Davassi A, Scopes G, Pirani A, Webster T, Brew F, Bloor C, Griffiths S, Bentley AR, Alda M, Jack P, Phillips AL, Edwards KJ. Characterization of a wheat breeders' Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum) Plant Biotechnology Journal. 2017;15:390–401. doi: 10.1111/pbi.12635. - DOI - PMC - PubMed
-
- Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O, Johnson R, Enk J, Periyannan S, Singh N, Asyraf Md Hatta M, Athiyannan N, Cheema J, Yu G, Kangara N, Ghosh S, Szabo LJ, Poland J, Bariana H, Jones JDG, Bentley AR, Ayliffe M, Olson E, Xu SS, Steffenson BJ, Lagudah E, Wulff BBH. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nature Biotechnology. 2019;37:139–143. doi: 10.1038/s41587-018-0007-9. - DOI - PubMed
-
- Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K, Jordan KW, Golan G, Deek J, Ben-Zvi B, Ben-Zvi G, Himmelbach A, MacLachlan RP, Sharpe AG, Fritz A, Ben-David R, Budak H, Fahima T, Korol A, Faris JD, Hernandez A, Mikel MA, Levy AA, Steffenson B, Maccaferri M, Tuberosa R, Cattivelli L, Faccioli P, Ceriotti A, Kashkush K, Pourkheirandish M, Komatsuda T, Eilam T, Sela H, Sharon A, Ohad N, Chamovitz DA, Mayer KFX, Stein N, Ronen G, Peleg Z, Pozniak CJ, Akhunov ED, Distelfeld A. Wild Emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 2017;357:93–97. doi: 10.1126/science.aan0032. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- RGS\R1\191163/Royal Society
- BB/M014045/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MBBIS 06-400258-12580/Monsanto-Beachell Borlaug International Scholarship
- BBS/E/J/000C0628/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011666/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P010741/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P013511/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M008908/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P016855/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

