RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML
- PMID: 32208489
- PMCID: PMC7094007
- DOI: 10.1182/bloodadvances.2019000901
RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML
Abstract
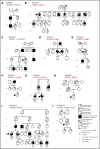
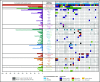
First reported in 1999, germline runt-related transcription factor 1 (RUNX1) mutations are a well-established cause of familial platelet disorder with predisposition to myeloid malignancy (FPD-MM). We present the clinical phenotypes and genetic mutations detected in 10 novel RUNX1-mutated FPD-MM families. Genomic analyses on these families detected 2 partial gene deletions, 3 novel mutations, and 5 recurrent mutations as the germline RUNX1 alterations leading to FPD-MM. Combining genomic data from the families reported herein with aggregated published data sets resulted in 130 germline RUNX1 families, which allowed us to investigate whether specific germline mutation characteristics (type, location) could explain the large phenotypic heterogeneity between patients with familial platelet disorder and different HMs. Comparing the somatic mutational signatures between the available familial (n = 35) and published sporadic (n = 137) RUNX1-mutated AML patients showed enrichment for somatic mutations affecting the second RUNX1 allele and GATA2. Conversely, we observed a decreased number of somatic mutations affecting NRAS, SRSF2, and DNMT3A and the collective genes associated with CHIP and epigenetic regulation. This is the largest aggregation and analysis of germline RUNX1 mutations performed to date, providing a unique opportunity to examine the factors underlying phenotypic differences and disease progression from FPD to MM.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures



References
-
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. - PubMed
-
- Song W-J, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23(2):166-175. - PubMed
-
- Brown AL, Churpek JE, Malcovati L, Döhner H, Godley LA. Recognition of familial myeloid neoplasia in adults. Semin Hematol. 2017;54(2):60-68. - PubMed
-
- Antony-Debré I, Duployez N, Bucci M, et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia. 2016;30(4):999-1002. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

