Space is the Place: Effects of Continuous Spatial Structure on Analysis of Population Genetic Data
- PMID: 32209569
- PMCID: PMC7198281
- DOI: 10.1534/genetics.120.303143
Space is the Place: Effects of Continuous Spatial Structure on Analysis of Population Genetic Data
Abstract
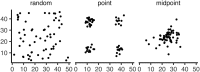
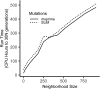
Real geography is continuous, but standard models in population genetics are based on discrete, well-mixed populations. As a result, many methods of analyzing genetic data assume that samples are a random draw from a well-mixed population, but are applied to clustered samples from populations that are structured clinally over space. Here, we use simulations of populations living in continuous geography to study the impacts of dispersal and sampling strategy on population genetic summary statistics, demographic inference, and genome-wide association studies (GWAS). We find that most common summary statistics have distributions that differ substantially from those seen in well-mixed populations, especially when Wright's neighborhood size is < 100 and sampling is spatially clustered. "Stepping-stone" models reproduce some of these effects, but discretizing the landscape introduces artifacts that in some cases are exacerbated at higher resolutions. The combination of low dispersal and clustered sampling causes demographic inference from the site frequency spectrum to infer more turbulent demographic histories, but averaged results across multiple simulations revealed surprisingly little systematic bias. We also show that the combination of spatially autocorrelated environments and limited dispersal causes GWAS to identify spurious signals of genetic association with purely environmentally determined phenotypes, and that this bias is only partially corrected by regressing out principal components of ancestry. Last, we discuss the relevance of our simulation results for inference from genetic variation in real organisms.
Keywords: GWAS; demography; haplotype block sharing; population structure; space.
Copyright © 2020 by the Genetics Society of America.
Figures







References
-
- Allee W. C., Park O., Emerson A. E., Park T., Schmidt K. P. et al. , 1949. Principles of Animal Ecology. Technical Report. Saunders Company, Philadelphia, PA.
-
- Antlfinger A. E., 1982. Genetic neighborhood structure of the salt marsh composite, Borrichia frutescens. J. Hered. 73: 128–132. 10.1093/oxfordjournals.jhered.a109595 - DOI
-
- Antolin M. F., Horne B. V., Berger M. D. Jr., Holloway A. K., Roach J. L. et al. , 2001. Effective population size and genetic structure of a piute ground squirrel (Spermophilus mollis) population. Can. J. Zool. 79: 26–34. 10.1139/z00-170 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

