Scale-invariant topology and bursty branching of evolutionary trees emerge from niche construction
- PMID: 32209672
- PMCID: PMC7149460
- DOI: 10.1073/pnas.1915088117
Scale-invariant topology and bursty branching of evolutionary trees emerge from niche construction
Abstract
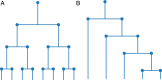
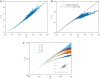
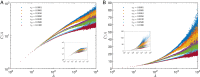
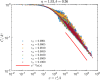
Phylogenetic trees describe both the evolutionary process and community diversity. Recent work has established that they exhibit scale-invariant topology, which quantifies the fact that their branching lies in between the two extreme cases of balanced binary trees and maximally unbalanced ones. In addition, the backbones of phylogenetic trees exhibit bursts of diversification on all timescales. Here, we present a simple, coarse-grained statistical model of niche construction coupled to speciation. Finite-size scaling analysis of the dynamics shows that the resultant phylogenetic tree topology is scale-invariant due to a singularity arising from large niche construction fluctuations that follow extinction events. The same model recapitulates the bursty pattern of diversification in time. These results show how dynamical scaling laws of phylogenetic trees on long timescales can reflect the indelible imprint of the interplay between ecological and evolutionary processes.
Keywords: evolution; molecular phylogeny; niche construction; scaling laws.
Figures
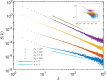
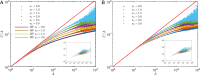
References
-
- Haig D., Hurst L. D., A quantitative measure of error minimization in the genetic code. J. Mol. Evol. 33, 412–417 (1991). - PubMed
-
- Koonin E. V., Novozhilov A. S., Origin and evolution of the universal genetic code. Annu. Rev. Genet. 51, 45–62 (2017). - PubMed
-
- Hernandez-Garcia E., Tuğrul M., Alejandro Herrada E., Eguiluz V. M., Klemm K., Simple models for scaling in phylogenetic trees. Int. J. Bifurcation Chaos 20, 805–811 (2010).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

