Tailoring a Global Iron Regulon to a Uropathogen
- PMID: 32209682
- PMCID: PMC7157518
- DOI: 10.1128/mBio.00351-20
Tailoring a Global Iron Regulon to a Uropathogen
Abstract
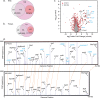
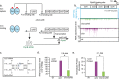
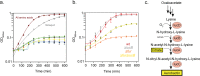
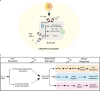
Pathogenicity islands and plasmids bear genes for pathogenesis of various Escherichia coli pathotypes. Although there is a basic understanding of the contribution of these virulence factors to disease, less is known about variation in regulatory networks in determining disease phenotypes. Here, we dissected a regulatory network directed by the conserved iron homeostasis regulator, ferric uptake regulator (Fur), in uropathogenic E. coli (UPEC) strain CFT073. Comparing anaerobic genome-scale Fur DNA binding with Fur-dependent transcript expression and protein levels of the uropathogen to that of commensal E. coli K-12 strain MG1655 showed that the Fur regulon of the core genome is conserved but also includes genes within the pathogenicity/genetic islands. Unexpectedly, regulons indicative of amino acid limitation and the general stress response were also indirectly activated in the uropathogen fur mutant, suggesting that induction of the Fur regulon increases amino acid demand. Using RpoS levels as a proxy, addition of amino acids mitigated the stress. In addition, iron chelation increased RpoS to the same levels as in the fur mutant. The increased amino acid demand of the fur mutant or iron chelated cells was exacerbated by aerobic conditions, which could be partly explained by the O2-dependent synthesis of the siderophore aerobactin, encoded by an operon within a pathogenicity island. Taken together, these data suggest that in the iron-poor environment of the urinary tract, amino acid availability could play a role in the proliferation of this uropathogen, particularly if there is sufficient O2 to produce aerobactin.IMPORTANCE Host iron restriction is a common mechanism for limiting the growth of pathogens. We compared the regulatory network controlled by Fur in uropathogenic E. coli (UPEC) to that of nonpathogenic E. coli K-12 to uncover strategies that pathogenic bacteria use to overcome iron limitation. Although iron homeostasis functions were regulated by Fur in the uropathogen as expected, a surprising finding was the activation of the stringent and general stress responses in the uropathogen fur mutant, which was rescued by amino acid addition. This coordinated global response could be important in controlling growth and survival under nutrient-limiting conditions and during transitions from the nutrient-rich environment of the lower gastrointestinal (GI) tract to the more restrictive environment of the urinary tract. The coupling of the response of iron limitation to increased demand for amino acids could be a critical attribute that sets UPEC apart from other E. coli pathotypes.
Keywords: CFT073; Fur; RyhB; Sigma S; UPEC; iron regulation; metabolic adaptation; ppGpp.
Copyright © 2020 Banerjee et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

