Divergent Evolution of Legionella RCC1 Repeat Effectors Defines the Range of Ran GTPase Cycle Targets
- PMID: 32209684
- PMCID: PMC7157520
- DOI: 10.1128/mBio.00405-20
Divergent Evolution of Legionella RCC1 Repeat Effectors Defines the Range of Ran GTPase Cycle Targets
Abstract
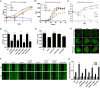
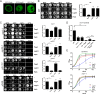
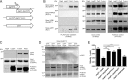
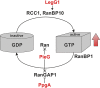
Legionella pneumophila governs its interactions with host cells by secreting >300 different "effector" proteins. Some of these effectors contain eukaryotic domains such as the RCC1 (regulator of chromosome condensation 1) repeats promoting the activation of the small GTPase Ran. In this report, we reveal a conserved pattern of L. pneumophila RCC1 repeat genes, which are distributed in two main clusters of strains. Accordingly, strain Philadelphia-1 contains two RCC1 genes implicated in bacterial virulence, legG1 (Legionella eukaryotic gene 1), and ppgA, while strain Paris contains only one, pieG The RCC1 repeat effectors localize to different cellular compartments and bind distinct components of the Ran GTPase cycle, including Ran modulators and the small GTPase itself, and yet they all promote the activation of Ran. The pieG gene spans the corresponding open reading frames of legG1 and a separate adjacent upstream gene, lpg1975legG1 and lpg1975 are fused upon addition of a single nucleotide to encode a protein that adopts the binding specificity of PieG. Thus, a point mutation in pieG splits the gene, altering the effector target. These results indicate that divergent evolution of RCC1 repeat effectors defines the Ran GTPase cycle targets and that modulation of different components of the cycle might fine-tune Ran activation during Legionella infection.IMPORTANCELegionella pneumophila is a ubiquitous environmental bacterium which, upon inhalation, causes a life-threatening pneumonia termed Legionnaires' disease. The opportunistic pathogen grows in amoebae and macrophages by employing a "type IV" secretion system, which secretes more than 300 different "effector" proteins into the host cell, where they subvert pivotal processes. The function of many of these effector proteins is unknown, and their evolution has not been studied. L. pneumophila RCC1 repeat effectors target the small GTPase Ran, a molecular switch implicated in different cellular processes such as nucleocytoplasmic transport and microtubule cytoskeleton dynamics. We provide evidence that one or more RCC1 repeat genes are distributed in two main clusters of L. pneumophila strains and have divergently evolved to target different components of the Ran GTPase activation cycle at different subcellular sites. Thus, L. pneumophila employs a sophisticated strategy to subvert host cell Ran GTPase during infection.
Keywords: Acanthamoeba; Dictyostelium; Legionella; amoeba; bacterial evolution; effector protein; guanine nucleotide exchange factor; host-pathogen interaction; macrophage; microtubule; pathogen vacuole; phosphoinositide lipid; small GTPase; type IV secretion; vesicle trafficking.
Copyright © 2020 Swart et al.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
