Completing Autophagy: Formation and Degradation of the Autophagic Body and Metabolite Salvage in Plants
- PMID: 32210003
- PMCID: PMC7139740
- DOI: 10.3390/ijms21062205
Completing Autophagy: Formation and Degradation of the Autophagic Body and Metabolite Salvage in Plants
Abstract
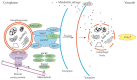
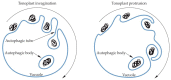
Autophagy is an evolutionarily conserved process that occurs in yeast, plants, and animals. Despite many years of research, some aspects of autophagy are still not fully explained. This mostly concerns the final stages of autophagy, which have not received as much interest from the scientific community as the initial stages of this process. The final stages of autophagy that we take into consideration in this review include the formation and degradation of the autophagic bodies as well as the efflux of metabolites from the vacuole to the cytoplasm. The autophagic bodies are formed through the fusion of an autophagosome and vacuole during macroautophagy and by vacuolar membrane invagination or protrusion during microautophagy. Then they are rapidly degraded by vacuolar lytic enzymes, and products of the degradation are reused. In this paper, we summarize the available information on the trafficking of the autophagosome towards the vacuole, the fusion of the autophagosome with the vacuole, the formation and decomposition of autophagic bodies inside the vacuole, and the efflux of metabolites to the cytoplasm. Special attention is given to the formation and degradation of autophagic bodies and metabolite salvage in plant cells.
Keywords: Atg proteins; SNARE proteins; autophagosome; tonoplast; vacuole.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The Atg17-Atg31-Atg29 complex and Atg11 regulate autophagosome-vacuole fusion.Autophagy. 2016 May 3;12(5):894-5. doi: 10.1080/15548627.2016.1162364. Autophagy. 2016. PMID: 26986547 Free PMC article. Review.
-
Dissection of autophagy in tobacco BY-2 cells under sucrose starvation conditions using the vacuolar H(+)-ATPase inhibitor concanamycin A and the autophagy-related protein Atg8.Plant Signal Behav. 2015;10(11):e1082699. doi: 10.1080/15592324.2015.1082699. Plant Signal Behav. 2015. PMID: 26368310 Free PMC article.
-
A method for the isolation and characterization of autophagic bodies from yeast provides a key tool to investigate cargos of autophagy.J Biol Chem. 2022 Dec;298(12):102641. doi: 10.1016/j.jbc.2022.102641. Epub 2022 Oct 25. J Biol Chem. 2022. PMID: 36306824 Free PMC article.
-
The Atg17-Atg31-Atg29 Complex Coordinates with Atg11 to Recruit the Vam7 SNARE and Mediate Autophagosome-Vacuole Fusion.Curr Biol. 2016 Jan 25;26(2):150-160. doi: 10.1016/j.cub.2015.11.054. Epub 2016 Jan 7. Curr Biol. 2016. PMID: 26774783 Free PMC article. Review.
-
Autophagosome Biogenesis and the Endoplasmic Reticulum: A Plant Perspective.Trends Plant Sci. 2018 Aug;23(8):677-692. doi: 10.1016/j.tplants.2018.05.002. Epub 2018 Jun 18. Trends Plant Sci. 2018. PMID: 29929776 Review.
Cited by
-
Plant Cell and Organism Development.Int J Mol Sci. 2020 Aug 6;21(16):5636. doi: 10.3390/ijms21165636. Int J Mol Sci. 2020. PMID: 32781648 Free PMC article.
-
Taro raphide-associated proteins: Allergens and crystal growth.Plant Direct. 2022 Sep 2;6(9):e443. doi: 10.1002/pld3.443. eCollection 2022 Sep. Plant Direct. 2022. PMID: 36091877 Free PMC article.
-
Autophagy: a necessary evil in cancer and inflammation.3 Biotech. 2024 Mar;14(3):87. doi: 10.1007/s13205-023-03864-w. Epub 2024 Feb 20. 3 Biotech. 2024. PMID: 38390576 Free PMC article. Review.
-
Regulation of autophagy by non-coding RNAs in human glioblastoma.Med Oncol. 2024 Oct 7;41(11):260. doi: 10.1007/s12032-024-02513-3. Med Oncol. 2024. PMID: 39375229 Review.
-
A View into Seed Autophagy: From Development to Environmental Responses.Plants (Basel). 2022 Nov 26;11(23):3247. doi: 10.3390/plants11233247. Plants (Basel). 2022. PMID: 36501287 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

