Evolutionary Trajectory for the Emergence of Novel Coronavirus SARS-CoV-2
- PMID: 32210130
- PMCID: PMC7157669
- DOI: 10.3390/pathogens9030240
Evolutionary Trajectory for the Emergence of Novel Coronavirus SARS-CoV-2
Abstract
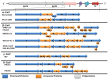
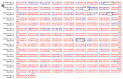
Over the last two decades, the world experienced three outbreaks of coronaviruses with elevated morbidity rates. Currently, the global community is facing emerging virus SARS-CoV-2 belonging to Betacoronavirus, which appears to be more transmissible but less deadly than SARS-CoV. The current study aimed to track the evolutionary ancestors and different evolutionary strategies that were genetically adapted by SARS-CoV-2. Our whole-genome analysis revealed that SARS-CoV-2 was the descendant of Bat SARS/SARS-like CoVs and bats served as a natural reservoir. SARS-CoV-2 used mutations and recombination as crucial strategies in different genomic regions including the envelop, membrane, nucleocapsid, and spike glycoproteins to become a novel infectious agent. We confirmed that mutations in different genomic regions of SARS-CoV-2 have specific influence on virus reproductive adaptability, allowing for genotype adjustment and adaptations in rapidly changing environments. Moreover, for the first time we identified nine putative recombination patterns in SARS-CoV-2, which encompass spike glycoprotein, RdRp, helicase and ORF3a. Six recombination regions were spotted in the S gene and are undoubtedly important for evolutionary survival, meanwhile this permitted the virus to modify superficial antigenicity to find a way from immune reconnaissance in animals and adapt to a human host. With these combined natural selected strategies, SARS-CoV-2 emerged as a novel virus in human society.
Keywords: SARS-CoV; SARS-CoV-2; evolutionary strategies; genomic structure; mutations; phylogeny; recombination or reassortment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gorbalenya E.A., Baker S.C., Baric R.S., Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. bioRxiv. 2020 doi: 10.1101/2020.02.07.937862. - DOI
-
- Masters P.S., Perlman S. Coronaviridae. In: Knipe D.M., Howley P.M., editors. Fields Virology. 6th ed. Lippincott Williams & Wilkins; New York, NY, USA: 2013. pp. 825–858.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

