Dimensionality reduction by UMAP to visualize physical and genetic interactions
- PMID: 32210240
- PMCID: PMC7093466
- DOI: 10.1038/s41467-020-15351-4
Dimensionality reduction by UMAP to visualize physical and genetic interactions
Abstract
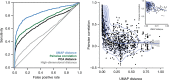
Dimensionality reduction is often used to visualize complex expression profiling data. Here, we use the Uniform Manifold Approximation and Projection (UMAP) method on published transcript profiles of 1484 single gene deletions of Saccharomyces cerevisiae. Proximity in low-dimensional UMAP space identifies groups of genes that correspond to protein complexes and pathways, and finds novel protein interactions, even within well-characterized complexes. This approach is more sensitive than previous methods and should be broadly useful as additional transcriptome datasets become available for other organisms.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hughes Timothy R, Marton Matthew J, Jones Allan R, Roberts Christopher J, Stoughton Roland, Armour Christopher D, Bennett Holly A, Coffey Ernest, Dai Hongyue, He Yudong D, Kidd Matthew J, King Amy M, Meyer Michael R, Slade David, Lum Pek Y, Stepaniants Sergey B, Shoemaker Daniel D, Gachotte Daniel, Chakraburtty Kalpana, Simon Julian, Bard Martin, Friend Stephen H. Functional Discovery via a Compendium of Expression Profiles. Cell. 2000;102(1):109–126. doi: 10.1016/S0092-8674(00)00015-5. - DOI - PubMed
-
- Laurens van der Maaten, Hinton G. Visualizing data using t-SNE Laurens. J. Mach. Learn. Res. 2008;9:2579–2605.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

