An African origin for Mycobacterium bovis
- PMID: 32211193
- PMCID: PMC7081938
- DOI: 10.1093/emph/eoaa005
An African origin for Mycobacterium bovis
Abstract
Background and objectives: Mycobacterium bovis and Mycobacterium caprae are two of the most important agents of tuberculosis in livestock and the most important causes of zoonotic tuberculosis in humans. However, little is known about the global population structure, phylogeography and evolutionary history of these pathogens.
Methodology: We compiled a global collection of 3364 whole-genome sequences from M.bovis and M.caprae originating from 35 countries and inferred their phylogenetic relationships, geographic origins and age.
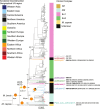
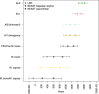
Results: Our results resolved the phylogenetic relationship among the four previously defined clonal complexes of M.bovis, and another eight newly described here. Our phylogeographic analysis showed that M.bovis likely originated in East Africa. While some groups remained restricted to East and West Africa, others have subsequently dispersed to different parts of the world.
Conclusions and implications: Our results allow a better understanding of the global population structure of M.bovis and its evolutionary history. This knowledge can be used to define better molecular markers for epidemiological investigations of M.bovis in settings where whole-genome sequencing cannot easily be implemented.
Lay summary: During the last few years, analyses of large globally representative collections of whole-genome sequences (WGS) from the human-adapted Mycobacterium tuberculosis complex (MTBC) lineages have enhanced our understanding of the global population structure, phylogeography and evolutionary history of these pathogens. In contrast, little corresponding data exists for M. bovis, the most important agent of tuberculosis in livestock. Using whole-genome sequences of globally distributed M. bovis isolates, we inferred the genetic relationships among different M. bovis genotypes distributed around the world. The most likely origin of M. bovis is East Africa according to our inferences. While some M. bovis groups remained restricted to East and West Africa, others have subsequently dispersed to different parts of the world driven by cattle movements.
Keywords: Mycobacterium bovis; bovine tuberculosis; molecular clock; phylogeography; whole-genome sequencing; zoonosis.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Foundation for Evolution, Medicine, and Public Health.
Figures

References
-
- WHO. Global Tuberculosis Report 2018. Geneva: World Health Organization, 2018.
-
- Olea-Popelka F, Muwonge A, Perera A. et al. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis—a call for action. Lancet Infect Dis 2017;17:e21–5. - PubMed
-
- Waters WR, Palmer MV, Buddle BM. et al. Bovine tuberculosis vaccine research: historical perspectives and recent advances. Vaccine 2012;30:2611–22. - PubMed
-
- Tschopp R, Hattendorf J, Roth F. et al. Cost estimate of bovine tuberculosis to Ethiopia. Curr Topics Microbiol Immunol 2013;365:249–68. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

