Epidemiological and Clinical Predictors of COVID-19
- PMID: 32211755
- PMCID: PMC7542554
- DOI: 10.1093/cid/ciaa322
Epidemiological and Clinical Predictors of COVID-19
Abstract
Background: Rapid identification of COVID-19 cases, which is crucial to outbreak containment efforts, is challenging due to the lack of pathognomonic symptoms and in settings with limited capacity for specialized nucleic acid-based reverse transcription polymerase chain reaction (PCR) testing.
Methods: This retrospective case-control study involves subjects (7-98 years) presenting at the designated national outbreak screening center and tertiary care hospital in Singapore for SARS-CoV-2 testing from 26 January to 16 February 2020. COVID-19 status was confirmed by PCR testing of sputum, nasopharyngeal swabs, or throat swabs. Demographic, clinical, laboratory, and exposure-risk variables ascertainable at presentation were analyzed to develop an algorithm for estimating the risk of COVID-19. Model development used Akaike's information criterion in a stepwise fashion to build logistic regression models, which were then translated into prediction scores. Performance was measured using receiver operating characteristic curves, adjusting for overconfidence using leave-one-out cross-validation.
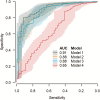
Results: The study population included 788 subjects, of whom 54 (6.9%) were SARS-CoV-2 positive and 734 (93.1%) were SARS-CoV-2 negative. The median age was 34 years, and 407 (51.7%) were female. Using leave-one-out cross-validation, all the models incorporating clinical tests (models 1, 2, and 3) performed well with areas under the receiver operating characteristic curve (AUCs) of 0.91, 0.88, and 0.88, respectively. In comparison, model 4 had an AUC of 0.65.
Conclusions: Rapidly ascertainable clinical and laboratory data could identify individuals at high risk of COVID-19 and enable prioritization of PCR testing and containment efforts. Basic laboratory test results were crucial to prediction models.
Keywords: COVID-19; SARS-CoV-2; prediction model; risk factors.
© The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures

Comment in
-
Good IgA Bad IgG in SARS-CoV-2 Infection?Clin Infect Dis. 2020 Jul 28;71(15):897-898. doi: 10.1093/cid/ciaa426. Clin Infect Dis. 2020. PMID: 32280952 Free PMC article. No abstract available.
References
-
- Wuhan Municipal Health Commission. Report of clustering pneumonia of unknown etiology in Wuhan City 2019. Available at: http://wjw.wuhan.gov.cn/front/web/showDetail/2019123108989. Accessed 19 February 2020.
-
- Zhu N, Zhang D, Wang W, et al. . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. Available at: https://doi.org/10.1056/NEJMoa2001017. Accessed 10 February 2020. - PMC - PubMed
-
- Gorbalenya AE, Baker SC, Baric RS, et al. . Severe acute respiratory syndrome-related coronavirus: the species and its viruses—a statement of the Coronavirus Study Group. Microbiology 2020. Available at: http://biorxiv.org/lookup/doi/10.1101/2020.02.07.937862. Accessed 19 February 2020. - DOI
-
- World Health Organization. Novel coronavirus (2019-nCoV) situation report—22 2020. Available at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/2.... Accessed 19 February 2020.
-
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020. Available at: https://jamanetwork.com/journals/jama/fullarticle/2762130. Accessed 3 March 2020. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

