The Drosophila Y Chromosome Affects Heterochromatin Integrity Genome-Wide
- PMID: 32211857
- PMCID: PMC7530609
- DOI: 10.1093/molbev/msaa082
The Drosophila Y Chromosome Affects Heterochromatin Integrity Genome-Wide
Abstract
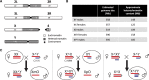
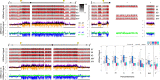
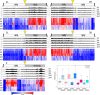
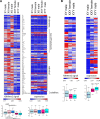
The Drosophila Y chromosome is gene poor and mainly consists of silenced, repetitive DNA. Nonetheless, the Y influences expression of hundreds of genes genome-wide, possibly by sequestering key components of the heterochromatin machinery away from other positions in the genome. To test the influence of the Y chromosome on the genome-wide chromatin landscape, we assayed the genomic distribution of histone modifications associated with gene activation (H3K4me3) or heterochromatin (H3K9me2 and H3K9me3) in fruit flies with varying sex chromosome complements (X0, XY, and XYY males; XX and XXY females). Consistent with the general deficiency of active chromatin modifications on the Y, we find that Y gene dose has little influence on the genomic distribution of H3K4me3. In contrast, both the presence and the number of Y chromosomes strongly influence genome-wide enrichment patterns of repressive chromatin modifications. Highly repetitive regions such as the pericentromeres, the dot, and the Y chromosome (if present) are enriched for heterochromatic modifications in wildtype males and females, and even more strongly in X0 flies. In contrast, the additional Y chromosome in XYY males and XXY females diminishes the heterochromatic signal in these normally silenced, repeat-rich regions, which is accompanied by an increase in expression of Y-linked repeats. We find hundreds of genes that are expressed differentially between individuals with aberrant sex chromosome karyotypes, many of which also show sex-biased expression in wildtype Drosophila. Thus, Y chromosomes influence heterochromatin integrity genome-wide, and differences in the chromatin landscape of males and females may also contribute to sex-biased gene expression and sexual dimorphisms.
Keywords: Drosophila; Y chromosome; chromatin sink; heterochromatin; sex chromosomes.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Ananiev EV, Barsky VE, Ilyin YV, Ryzic MV.. 1984. The arrangement of transposable elements in the polytene chromosomes of Drosophila melanogaster. Chromosoma 90(5):366–377.
-
- Baker WK. 1968. Position-effect variegation In: Caspari EW, editor. Advances in genetics. Academic Press; p. 133–169. - PubMed
-
- Belyaeva ES, Demakova OV, Umbetova GH, Zhimulev IF.. 1993. Cytogenetic and molecular aspects of position-effect variegation in Drosophila melanogaster. V. Heterochromatin-associated protein HP1 appears in euchromatic chromosomal regions that are inactivated as a result of position-effect variegation. Chromosoma 102(8):583–590. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

