Heparan sulfate is a clearance receptor for aberrant extracellular proteins
- PMID: 32211892
- PMCID: PMC7054991
- DOI: 10.1083/jcb.201911126
Heparan sulfate is a clearance receptor for aberrant extracellular proteins
Abstract
The accumulation of aberrant proteins leads to various neurodegenerative disorders. Mammalian cells contain several intracellular protein degradation systems, including autophagy and proteasomal systems, that selectively remove aberrant intracellular proteins. Although mammals contain not only intracellular but also extracellular proteins, the mechanism underlying the quality control of aberrant extracellular proteins is poorly understood. Here, using a novel quantitative fluorescence assay and genome-wide CRISPR screening, we identified the receptor-mediated degradation pathway by which misfolded extracellular proteins are selectively captured by the extracellular chaperone Clusterin and undergo endocytosis via the cell surface heparan sulfate (HS) receptor. Biochemical analyses revealed that positively charged residues on Clusterin electrostatically interact with negatively charged HS. Furthermore, the Clusterin-HS pathway facilitates the degradation of amyloid β peptide and diverse leaked cytosolic proteins in extracellular space. Our results identify a novel protein quality control system for preserving extracellular proteostasis and highlight its role in preventing diseases associated with aberrant extracellular proteins.
© 2020 Itakura et al.
Figures
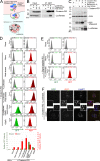

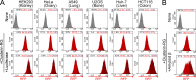
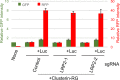





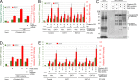
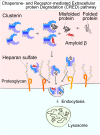
Comment in
-
Heparan sulfate and clusterin: Cleaning squad for extracellular protein degradation.J Cell Biol. 2020 Mar 2;219(3):e202001159. doi: 10.1083/jcb.202001159. J Cell Biol. 2020. PMID: 32211896 Free PMC article.
References
-
- Bell R.D., Sagare A.P., Friedman A.E., Bedi G.S., Holtzman D.M., Deane R., and Zlokovic B.V.. 2007. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J. Cereb. Blood Flow Metab. 27:909–918. 10.1038/sj.jcbfm.9600419 - DOI - PMC - PubMed
-
- Bettens K., Brouwers N., Engelborghs S., Lambert J.C., Rogaeva E., Vandenberghe R., Le Bastard N., Pasquier F., Vermeulen S., Van Dongen J., et al. 2012. Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk. Mol. Neurodegener. 7:3 10.1186/1750-1326-7-3 - DOI - PMC - PubMed
-
- Boland B., Yu W.H., Corti O., Mollereau B., Henriques A., Bezard E., Pastores G.M., Rubinsztein D.C., Nixon R.A., Duchen M.R., et al. 2018. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 17:660–688. 10.1038/nrd.2018.109 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

