NeAT: a Nonlinear Analysis Toolbox for Neuroimaging
- PMID: 32212063
- PMCID: PMC7498484
- DOI: 10.1007/s12021-020-09456-w
NeAT: a Nonlinear Analysis Toolbox for Neuroimaging
Abstract
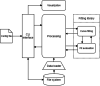
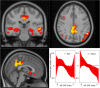
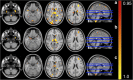
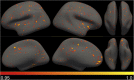
NeAT is a modular, flexible and user-friendly neuroimaging analysis toolbox for modeling linear and nonlinear effects overcoming the limitations of the standard neuroimaging methods which are solely based on linear models. NeAT provides a wide range of statistical and machine learning non-linear methods for model estimation, several metrics based on curve fitting and complexity for model inference and a graphical user interface (GUI) for visualization of results. We illustrate its usefulness on two study cases where non-linear effects have been previously established. Firstly, we study the nonlinear effects of Alzheimer's disease on brain morphology (volume and cortical thickness). Secondly, we analyze the effect of the apolipoprotein APOE-ε4 genotype on brain aging and its interaction with age. NeAT is fully documented and publicly distributed at https://imatge-upc.github.io/neat-tool/ .
Keywords: APOE; Alzheimer's disease; GAM; GLM; SVR; inference; neuroimaging; nonlinear.
Conflict of interest statement
Author Santi Puch is employed by company QMENTA and author Asier Aduriz is employed by company VILYNX. All other authors declare no competing interests.
Figures





References
-
- Breiman L, Friedman JH. Estimating optimal transformations for multiple regression and correlation. J Am Stat Assoc. 1985;80(391):580–598. doi: 10.1080/01621459.1985.10478157. - DOI
-
- Brett, M., Hanke, M., Cipollini, B., Côté, M. A., Markiewicz, C., Gerhard, S., Larson, E., Lee, G. R., Halchenko, Y., Kastman, E., Morency, F. C., Millman, J., Rokem, A., Gramfort, A., van den Bosch, J. J. F., Subramaniam, K., Nichols, N., Oosterhof, N. N., St-Jean, S., Amirbekian, B., Nimmo-Smith, I., Ghosh, S., Varoquaux, G., Garyfallidis, E. (2016). nibabel: 2.1. 0.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

