Cell wall/vacuolar inhibitor of fructosidase 1 regulates ABA response and salt tolerance in Arabidopsis
- PMID: 32213123
- PMCID: PMC7194370
- DOI: 10.1080/15592324.2020.1744293
Cell wall/vacuolar inhibitor of fructosidase 1 regulates ABA response and salt tolerance in Arabidopsis
Abstract
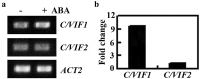
ABA regulates abiotic stress tolerance in plants via activating/repressing gene expression. However, the functions of many ABA response genes remained unknown. C/VIFs are proteinaceous inhibitors of the CWI and VI invertases. We report here the involvement of C/VIF1 in regulating ABA response and salt tolerance in Arabidopsis. We found that the expression level of C/VIF1 was increased in response to ABA treatment. By using CRISPR/Cas9 gene editing, we generated transgene-free c/vif1 mutants. We also generated C/VIF1 overexpression plants by expressing C/VIF1 under the control of the 35S promoter. We examined ABA response of the 35S:C/VIF1 transgenic plants and the c/vif1 mutants by using seed germination and seedling greening assays, and found that the 35S:C/VIF1 transgenic plants showed an enhanced sensitivity to ABA treatment in both assays. On the other hand, the c/vif1 mutants showed slight enhanced tolerance to ABA only at the early stage of germination. We also found that salt tolerance was reduced in the 35S:C/VIF1 transgenic plants in seed germination assays, but slightly increased in the c/vif1 mutants. Taken together, our results suggest that C/VIF1 is an ABA response gene, and C/VIF1 is involved in the regulation of ABA response and salt tolerance in Arabidopsis.
Keywords: Arabidopsis; Cell wall/vacuolar inhibitor of fructosidase 1; abscisic acid; gene editing; salt tolerance.
Figures




References
-
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K.. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Bio. 2006;9:436–442. doi:10.1016/j.pbi.2006.05.014. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
