Trans-Acting Small RNAs and Their Effects on Gene Expression in Escherichia coli and Salmonella enterica
- PMID: 32213244
- PMCID: PMC7112153
- DOI: 10.1128/ecosalplus.ESP-0030-2019
Trans-Acting Small RNAs and Their Effects on Gene Expression in Escherichia coli and Salmonella enterica
Abstract
The last few decades have led to an explosion in our understanding of the major roles that small regulatory RNAs (sRNAs) play in regulatory circuits and the responses to stress in many bacterial species. Much of the foundational work was carried out with Escherichia coli and Salmonella enterica serovar Typhimurium. The studies of these organisms provided an overview of how the sRNAs function and their impact on bacterial physiology, serving as a blueprint for sRNA biology in many other prokaryotes. They also led to the development of new technologies. In this chapter, we first summarize how these sRNAs were identified, defining them in the process. We discuss how they are regulated and how they act and provide selected examples of their roles in regulatory circuits and the consequences of this regulation. Throughout, we summarize the methodologies that were developed to identify and study the regulatory RNAs, most of which are applicable to other bacteria. Newly updated databases of the known sRNAs in E. coli K-12 and S. enterica Typhimurium SL1344 serve as a reference point for much of the discussion and, hopefully, as a resource for readers and for future experiments to address open questions raised in this review.
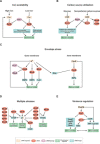
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

