Whole-genome sequencing of Burkholderia pseudomallei from an urban melioidosis hot spot reveals a fine-scale population structure and localised spatial clustering in the environment
- PMID: 32214186
- PMCID: PMC7096523
- DOI: 10.1038/s41598-020-62300-8
Whole-genome sequencing of Burkholderia pseudomallei from an urban melioidosis hot spot reveals a fine-scale population structure and localised spatial clustering in the environment
Abstract
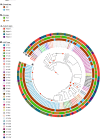
Melioidosis is a severe disease caused by the environmental bacterium Burkholderia pseudomallei that affects both humans and animals throughout northern Australia, Southeast Asia and increasingly globally. While there is a considerable degree of genetic diversity amongst isolates, B. pseudomallei has a robust global biogeographic structure and genetic populations are spatially clustered in the environment. We examined the distribution and local spread of B. pseudomallei in Darwin, Northern Territory, Australia, which has the highest recorded urban incidence of melioidosis globally. We sampled soil and land runoff throughout the city centre and performed whole-genome sequencing (WGS) on B. pseudomallei isolates. By combining phylogenetic analyses, Bayesian clustering and spatial hot spot analysis our results demonstrate that some sequence types (STs) are widespread in the urban Darwin environment, while others are highly spatially clustered over a small geographic scale. This clustering matches the spatial distribution of clinical cases for one ST. Results also demonstrate a greater overall isolate diversity recovered from drains compared to park soils, further supporting the role drains may play in dispersal of B. pseudomallei STs in the environment. Collectively, knowledge gained from this study will allow for better understanding of B. pseudomallei phylogeography and melioidosis source attribution, particularly on a local level.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

