A single amino acid substitution uncouples catalysis and allostery in an essential biosynthetic enzyme in Mycobacterium tuberculosis
- PMID: 32217694
- PMCID: PMC7212630
- DOI: 10.1074/jbc.RA120.012605
A single amino acid substitution uncouples catalysis and allostery in an essential biosynthetic enzyme in Mycobacterium tuberculosis
Abstract
Allostery exploits the conformational dynamics of enzymes by triggering a shift in population ensembles toward functionally distinct conformational or dynamic states. Allostery extensively regulates the activities of key enzymes within biosynthetic pathways to meet metabolic demand for their end products. Here, we have examined a critical enzyme, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAH7PS), at the gateway to aromatic amino acid biosynthesis in Mycobacterium tuberculosis, which shows extremely complex dynamic allostery: three distinct aromatic amino acids jointly communicate occupancy to the active site via subtle changes in dynamics, enabling exquisite fine-tuning of delivery of these essential metabolites. Furthermore, this allosteric mechanism is co-opted by pathway branchpoint enzyme chorismate mutase upon complex formation. In this study, using statistical coupling analysis, site-directed mutagenesis, isothermal calorimetry, small-angle X-ray scattering, and X-ray crystallography analyses, we have pinpointed a critical node within the complex dynamic communication network responsible for this sophisticated allosteric machinery. Through a facile Gly to Pro substitution, we have altered backbone dynamics, completely severing the allosteric signal yet remarkably, generating a nonallosteric enzyme that retains full catalytic activity. We also identified a second residue of prime importance to the inter-enzyme communication with chorismate mutase. Our results reveal that highly complex dynamic allostery is surprisingly vulnerable and provide further insights into the intimate link between catalysis and allostery.
Keywords: DAH7PS; allosteric regulation; aromatic amino acid biosynthesis; enzyme inhibitor; enzyme mutation; enzyme structure; molecular dynamics; protein sequence; shikimate pathway; statistical coupling analysis; tuberculosis.
© 2020 Jiao et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


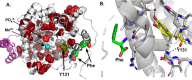
References
-
- Walker G. E., Dunbar B., Hunter I. S., Nimmo H. G., and Coggins J. R. (1996) Evidence for a novel class of microbial 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase in Streptomyces coelicolor A3(2), Streptomyces rimosus and Neurospora crassa. Microbiology 142, 1973–1982 - PubMed
-
- Wagner T., Shumilin I. A., Bauerle R., and Kretsinger R. H. (2000) Structure of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli: comparison of the Mn(2+)*2-phosphoglycolate and the Pb(2+)*2-phosphoenolpyruvate complexes and implications for catalysis. J. Mol. Biol. 301, 389–399 10.1006/jmbi.2000.3957 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

