VIRMA-Dependent N6-Methyladenosine Modifications Regulate the Expression of Long Non-Coding RNAs CCAT1 and CCAT2 in Prostate Cancer
- PMID: 32218194
- PMCID: PMC7226055
- DOI: 10.3390/cancers12040771
VIRMA-Dependent N6-Methyladenosine Modifications Regulate the Expression of Long Non-Coding RNAs CCAT1 and CCAT2 in Prostate Cancer
Abstract
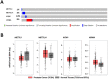
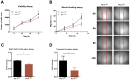
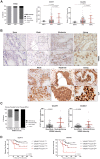
RNA methylation at position N6 in adenosine (m6A) and its associated methyltransferase complex (MTC) are involved in tumorigenesis. We aimed to explore m6A biological function for long non-coding RNAs (lncRNAs) in prostate cancer (PCa) and its clinical significance. m6A and MTC levels in PCa cells were characterized by ELISA and western blot. Putative m6A-regulated lncRNAs were identified and validated by lncRNA profiler qPCR array and bioinformatics analysis, followed by m6A/RNA co-immunoprecipitation. Impact of m6A depletion on RNA stability was assessed by Actinomycin D assay. The association of m6A-levels with PCa prognosis was examined in clinical samples. Higher m6A-levels and VIRMA overexpression were detected in metastatic castration-resistant PCa (mCRPC) cells (p < 0.05). VIRMA knockdown in PC-3 cells significantly decreased m6A-levels (p = 0.0317), attenuated malignant phenotype and suppressed the expression of oncogenic lncRNAs CCAT1 and CCAT2 (p < 0.00001). VIRMA depletion and m6A reduction decreased the stability and abundance of CCAT1/2 transcripts. Higher expression of VIRMA, CCAT1, and CCAT2 as a group variable was an independent predictor of poor prognosis (HR = 9.083, CI95% 1.911-43.183, p = 0.006). VIRMA is a critical factor sustaining m6A-levels in PCa cells. VIRMA downregulation attenuates the aggressive phenotype of PCa by overall reduction of m6A-levels decreasing stability and abundance of oncogenic lncRNAs.
Keywords: N6-Methyladenosine; VIRMA; epitranscriptome; long non-coding RNAs; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Esteller M., Pandolfi P.P. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 2017;7:359–368. doi: 10.1158/2159-8290.CD-16-1292. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

