Artificial Intelligence and Machine learning based prediction of resistant and susceptible mutations in Mycobacterium tuberculosis
- PMID: 32218465
- PMCID: PMC7099008
- DOI: 10.1038/s41598-020-62368-2
Artificial Intelligence and Machine learning based prediction of resistant and susceptible mutations in Mycobacterium tuberculosis
Erratum in
-
Author Correction: Artificial Intelligence and Machine learning based prediction of resistant and susceptible mutations in Mycobacterium tuberculosis.Sci Rep. 2020 Sep 1;10(1):14660. doi: 10.1038/s41598-020-71840-y. Sci Rep. 2020. PMID: 32868840 Free PMC article.
Abstract
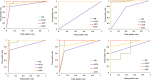
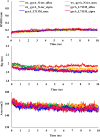
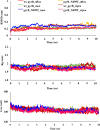
Tuberculosis (TB), an infectious disease caused by Mycobacterium tuberculosis (M.tb), causes highest number of deaths globally for any bacterial disease necessitating novel diagnosis and treatment strategies. High-throughput sequencing methods generate a large amount of data which could be exploited in determining multi-drug resistant (MDR-TB) associated mutations. The present work is a computational framework that uses artificial intelligence (AI) based machine learning (ML) approaches for predicting resistance in the genes rpoB, inhA, katG, pncA, gyrA and gyrB for the drugs rifampicin, isoniazid, pyrazinamide and fluoroquinolones. The single nucleotide variations were represented by several sequence and structural features that indicate the influence of mutations on the target protein coded by each gene. We used ML algorithms - naïve bayes, k nearest neighbor, support vector machine, and artificial neural network, to build the prediction models. The classification models had an average accuracy of 85% across all examined genes and were evaluated on an external unseen dataset to demonstrate their application. Further, molecular docking and molecular dynamics simulations were performed for wild type and predicted resistance causing mutant protein and anti-TB drug complexes to study their impact on the conformation of proteins to confirm the observed phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Organization, W. H. Global tuberculosis report, https://www.who.int/news-room/fact-sheets/detail/tuberculosis (2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

