Dynamic Transcriptional Responses to Injury of Regenerative and Non-regenerative Cardiomyocytes Revealed by Single-Nucleus RNA Sequencing
- PMID: 32220304
- PMCID: PMC7365574
- DOI: 10.1016/j.devcel.2020.02.019
Dynamic Transcriptional Responses to Injury of Regenerative and Non-regenerative Cardiomyocytes Revealed by Single-Nucleus RNA Sequencing
Erratum in
-
Dynamic Transcriptional Responses to Injury of Regenerative and Non-regenerative Cardiomyocytes Revealed by Single-Nucleus RNA Sequencing.Dev Cell. 2020 Dec 7;55(5):665-667. doi: 10.1016/j.devcel.2020.11.006. Dev Cell. 2020. PMID: 33290696 Free PMC article. No abstract available.
Abstract
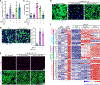
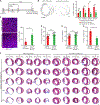
The adult mammalian heart is incapable of regeneration following injury. In contrast, the neonatal mouse heart can efficiently regenerate during the first week of life. The molecular mechanisms that mediate the regenerative response and its blockade in later life are not understood. Here, by single-nucleus RNA sequencing, we map the dynamic transcriptional landscape of five distinct cardiomyocyte populations in healthy, injured, and regenerating mouse hearts. We identify immature cardiomyocytes that enter the cell cycle following injury and disappear as the heart loses the ability to regenerate. These proliferative neonatal cardiomyocytes display a unique transcriptional program dependent on nuclear transcription factor Y subunit alpha (NFYa) and nuclear factor erythroid 2-like 1 (NFE2L1) transcription factors, which exert proliferative and protective functions, respectively. Cardiac overexpression of these two factors conferred protection against ischemic injury in mature mouse hearts that were otherwise non-regenerative. These findings advance our understanding of the cellular basis of neonatal heart regeneration and reveal a transcriptional landscape for heart repair following injury.
Keywords: NFE2L1; NFYa; cell survival; heart regeneration; ischemia; transcriptional response to injury.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




References
-
- Alkass K, Panula J, Westman M, Wu TD, Guerquin-Kern JL, and Bergmann O (2015). No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell 163, 1026–1036. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

