Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA
- PMID: 32220646
- PMCID: PMC7214211
- DOI: 10.1016/j.molcel.2020.02.016
Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA
Abstract
Metazoan microRNAs require specific maturation steps initiated by Microprocessor, comprising Drosha and DGCR8. Lack of structural information for the assembled complex has hindered an understanding of how Microprocessor recognizes primary microRNA transcripts (pri-miRNAs). Here we present a cryoelectron microscopy structure of human Microprocessor with a pri-miRNA docked in the active site, poised for cleavage. The basal junction is recognized by a four-way intramolecular junction in Drosha, triggered by the Belt and Wedge regions that clamp over the ssRNA. The belt is important for efficiency and accuracy of pri-miRNA processing. Two dsRBDs form a molecular ruler to measure the stem length between the two dsRNA-ssRNA junctions. The specific organization of the dsRBDs near the apical junction is independent of Drosha core domains, as observed in a second structure in the partially docked state. Collectively, we derive a molecular model to explain how Microprocessor recognizes a pri-miRNA and accurately identifies the cleavage site.
Keywords: DGCR8; Drosha; Microprocessor; RNA processing; RNase III; cryo-EM; dsRBD; microRNA; pri-miRNA; structure.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
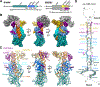
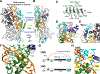




References
-
- Bernstein E, Caudy AA, Hammond SM, and Hannon GJ (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

