Microbial Residents of the Atlantis Massif's Shallow Serpentinite Subsurface
- PMID: 32220840
- PMCID: PMC7237769
- DOI: 10.1128/AEM.00356-20
Microbial Residents of the Atlantis Massif's Shallow Serpentinite Subsurface
Abstract
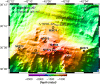
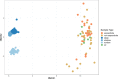
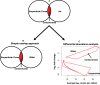
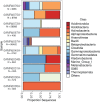
The Atlantis Massif rises 4,000 m above the seafloor near the Mid-Atlantic Ridge and consists of rocks uplifted from Earth's lower crust and upper mantle. Exposure of the mantle rocks to seawater leads to their alteration into serpentinites. These aqueous geochemical reactions, collectively known as the process of serpentinization, are exothermic and are associated with the release of hydrogen gas (H2), methane (CH4), and small organic molecules. The biological consequences of this flux of energy and organic compounds from the Atlantis Massif were explored by International Ocean Discovery Program (IODP) Expedition 357, which used seabed drills to collect continuous sequences of shallow (<16 m below seafloor) marine serpentinites and mafic assemblages. Here, we report the census of microbial diversity in samples of the drill cores, as measured by environmental 16S rRNA gene amplicon sequencing. The problem of contamination of subsurface samples was a primary concern during all stages of this project, starting from the initial study design, continuing to the collection of samples from the seafloor, handling the samples shipboard and in the lab, preparing the samples for DNA extraction, and analyzing the DNA sequence data. To distinguish endemic microbial taxa of serpentinite subsurface rocks from seawater residents and other potential contaminants, the distributions of individual 16S rRNA gene sequences among all samples were evaluated, taking into consideration both presence/absence and relative abundances. Our results highlight a few candidate residents of the shallow serpentinite subsurface, including uncultured representatives of the Thermoplasmata, Acidobacteria, Acidimicrobia, and ChloroflexiIMPORTANCE The International Ocean Discovery Program Expedition 357-"Serpentinization and Life"-utilized seabed drills to collect rocks from the oceanic crust. The recovered rock cores represent the shallow serpentinite subsurface of the Atlantis Massif, where reactions between uplifted mantle rocks and water, collectively known as serpentinization, produce environmental conditions that can stimulate biological activity and are thought to be analogous to environments that were prevalent on the early Earth and perhaps other planets. The methodology and results of this project have implications for life detection experiments, including sample return missions, and provide a window into the diversity of microbial communities inhabiting subseafloor serpentinites.
Keywords: Atlantis Massif; contamination; serpentinization.
Copyright © 2020 Motamedi et al.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

